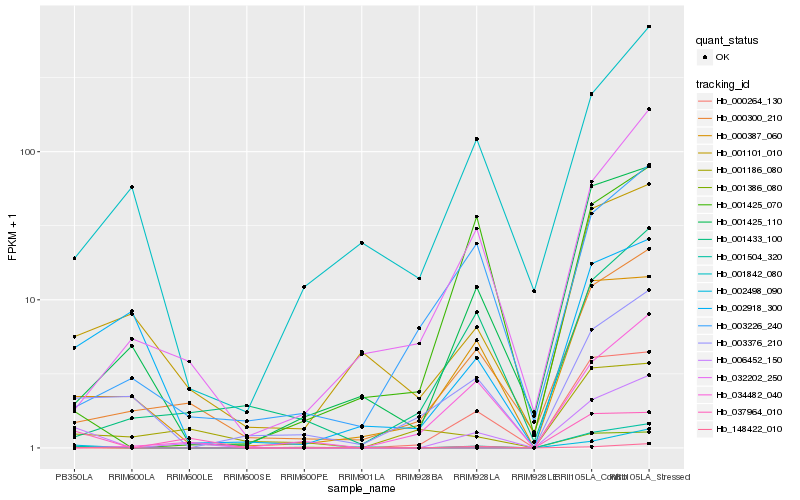
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006452_150 |
0.0 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor 7-like [Jatropha curcas] |
| 2 |
Hb_001504_320 |
0.2081296366 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 3 |
Hb_000300_210 |
0.2307782017 |
- |
- |
gd2b, putative [Ricinus communis] |
| 4 |
Hb_002498_090 |
0.233731585 |
- |
- |
- |
| 5 |
Hb_000387_060 |
0.2495419615 |
- |
- |
- |
| 6 |
Hb_001101_010 |
0.2547739759 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 7 |
Hb_001425_110 |
0.26089553 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 8 |
Hb_037964_010 |
0.2643087014 |
- |
- |
hypothetical protein MIMGU_mgv1a000906mg [Erythranthe guttata] |
| 9 |
Hb_003376_210 |
0.265527877 |
- |
- |
- |
| 10 |
Hb_032202_250 |
0.270234124 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 11 |
Hb_000264_130 |
0.2714529525 |
- |
- |
PREDICTED: uncharacterized protein LOC103870402 [Brassica rapa] |
| 12 |
Hb_001433_100 |
0.2756565619 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_034482_040 |
0.2811105821 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 14 |
Hb_003226_240 |
0.2853162702 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 15 |
Hb_001186_080 |
0.2917822488 |
- |
- |
PREDICTED: F-box protein FBW2-like [Nelumbo nucifera] |
| 16 |
Hb_001386_080 |
0.2918172086 |
- |
- |
hypothetical protein JCGZ_06052 [Jatropha curcas] |
| 17 |
Hb_148422_010 |
0.2923088623 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_001425_070 |
0.2924216768 |
- |
- |
- |
| 19 |
Hb_001842_080 |
0.2924823569 |
- |
- |
- |
| 20 |
Hb_002918_300 |
0.2927854808 |
- |
- |
- |