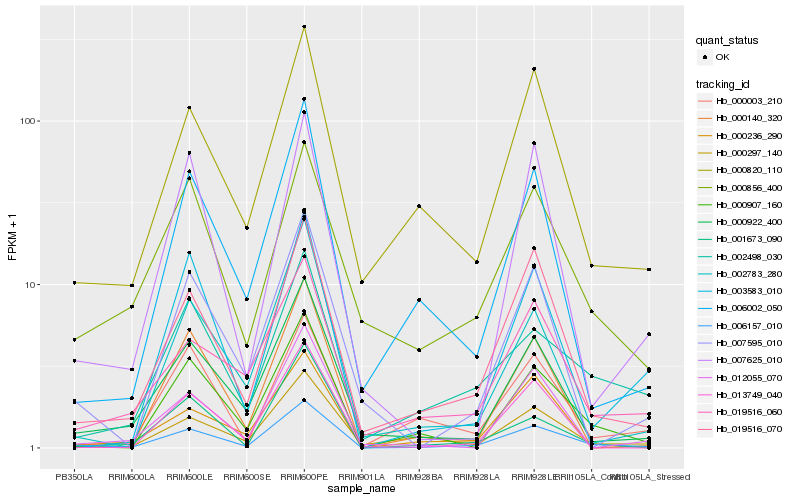
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006157_010 |
0.0 |
- |
- |
PREDICTED: endoglucanase 5 [Jatropha curcas] |
| 2 |
Hb_019516_070 |
0.1307113905 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 3 |
Hb_000922_400 |
0.1522313273 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 4 |
Hb_000820_110 |
0.1594599479 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 5 |
Hb_006002_050 |
0.1668021849 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 6 |
Hb_000907_160 |
0.1714818911 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 7 |
Hb_019516_060 |
0.1740905592 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 8 |
Hb_002783_280 |
0.1753482935 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 9 |
Hb_002498_030 |
0.1766290663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001673_090 |
0.1775947742 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 11 |
Hb_000297_140 |
0.179781392 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 12 |
Hb_007595_010 |
0.1803472343 |
- |
- |
Photosystem II reaction center protein K [Medicago truncatula] |
| 13 |
Hb_003583_010 |
0.1816023341 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 14 |
Hb_000003_210 |
0.1816251728 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 15 |
Hb_000140_320 |
0.1818829922 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 16 |
Hb_012055_070 |
0.1841683476 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_000236_290 |
0.1865276191 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 18 |
Hb_013749_040 |
0.1865803129 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PXL1 [Jatropha curcas] |
| 19 |
Hb_000856_400 |
0.1870725024 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 20 |
Hb_007625_010 |
0.1888176232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |