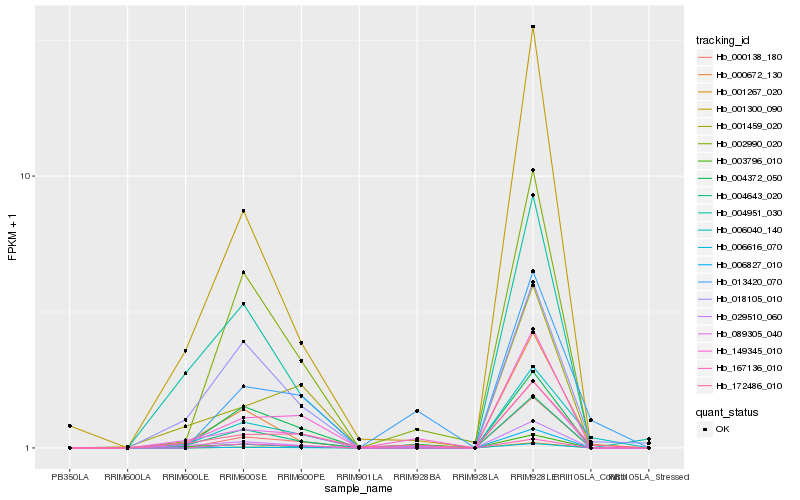
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006040_140 |
0.0 |
- |
- |
hypothetical protein NitaMp105 [Nicotiana tabacum] |
| 2 |
Hb_000672_130 |
0.1725790121 |
- |
- |
hypothetical protein POPTR_0010s22870g [Populus trichocarpa] |
| 3 |
Hb_172486_010 |
0.1730709761 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_004643_020 |
0.1846106876 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 5 |
Hb_018105_010 |
0.1945432909 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 6 |
Hb_089305_040 |
0.2001503854 |
- |
- |
- |
| 7 |
Hb_003796_010 |
0.2031179436 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 8 |
Hb_000138_180 |
0.2122824555 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 9 |
Hb_001300_090 |
0.2168924269 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 10 |
Hb_006827_010 |
0.2187941066 |
- |
- |
retrotransposon protein, putative, unclassified, expressed [Oryza sativa Japonica Group] |
| 11 |
Hb_013420_070 |
0.2195741433 |
- |
- |
hypothetical protein EUGRSUZ_I01852 [Eucalyptus grandis] |
| 12 |
Hb_002990_020 |
0.2198559541 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 13 |
Hb_001267_020 |
0.2210674081 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 14 |
Hb_004951_030 |
0.2295507372 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 15 |
Hb_029510_060 |
0.2303580016 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 16 |
Hb_006616_070 |
0.2310070177 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_149345_010 |
0.2341534643 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 18 |
Hb_004372_050 |
0.2359747291 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 19 |
Hb_001459_020 |
0.2363240477 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_167136_010 |
0.236574695 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |