| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
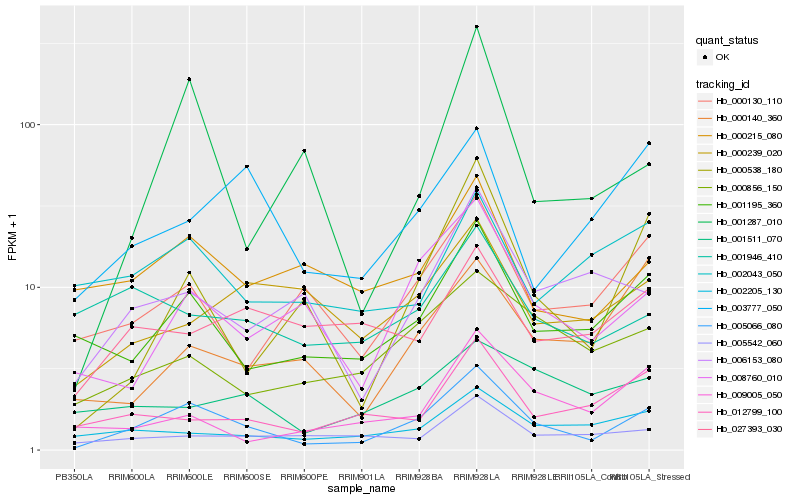
Hb_005066_080 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa025081mg [Prunus persica] |
| 2 |
Hb_001195_360 |
0.2030703692 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 3 |
Hb_008760_010 |
0.2149411671 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 4 |
Hb_000538_180 |
0.2294876648 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 5 |
Hb_000856_150 |
0.233922266 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 6 |
Hb_000130_110 |
0.2340975425 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 7 |
Hb_000215_080 |
0.2367744867 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006153_080 |
0.2388831043 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 9 |
Hb_002205_130 |
0.239775821 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 10 |
Hb_005542_060 |
0.244484595 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_012799_100 |
0.2492438305 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 12 |
Hb_009005_050 |
0.2542998332 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 13 |
Hb_001287_010 |
0.2544210072 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 14 |
Hb_003777_050 |
0.2545143249 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 15 |
Hb_001511_070 |
0.2547210537 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 16 |
Hb_000239_020 |
0.2556464098 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 17 |
Hb_001946_410 |
0.2561699554 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002043_050 |
0.2583620474 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 19 |
Hb_000140_360 |
0.2586486693 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 20 |
Hb_027393_030 |
0.2598099198 |
- |
- |
PREDICTED: uncharacterized protein At5g39865 [Jatropha curcas] |