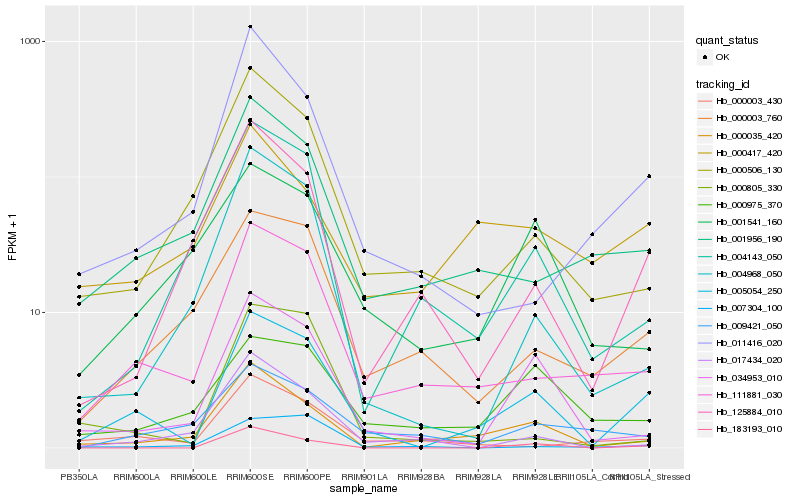
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280 [Jatropha curcas] |
| 2 |
Hb_111881_030 |
0.2164401843 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 3 |
Hb_034953_010 |
0.2298539683 |
- |
- |
hypothetical protein JCGZ_00031 [Jatropha curcas] |
| 4 |
Hb_000035_420 |
0.2406961644 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 5 |
Hb_183193_010 |
0.251700179 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 6 |
Hb_001956_190 |
0.2536883878 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 7 |
Hb_000003_760 |
0.2576839249 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 8 |
Hb_000003_430 |
0.2660475479 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000975_370 |
0.2743136842 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000506_130 |
0.274526209 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 11 |
Hb_011416_020 |
0.2749545684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004968_050 |
0.2750759847 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 13 |
Hb_125884_010 |
0.2768482104 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Jatropha curcas] |
| 14 |
Hb_004143_050 |
0.2774979714 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 15 |
Hb_001541_160 |
0.2810776385 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 16 |
Hb_000805_330 |
0.283351357 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 17 |
Hb_009421_050 |
0.2864622826 |
- |
- |
PREDICTED: uncharacterized protein LOC105647037 [Jatropha curcas] |
| 18 |
Hb_000417_420 |
0.2869451877 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 19 |
Hb_017434_020 |
0.2878356843 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 20 |
Hb_007304_100 |
0.2914087135 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |