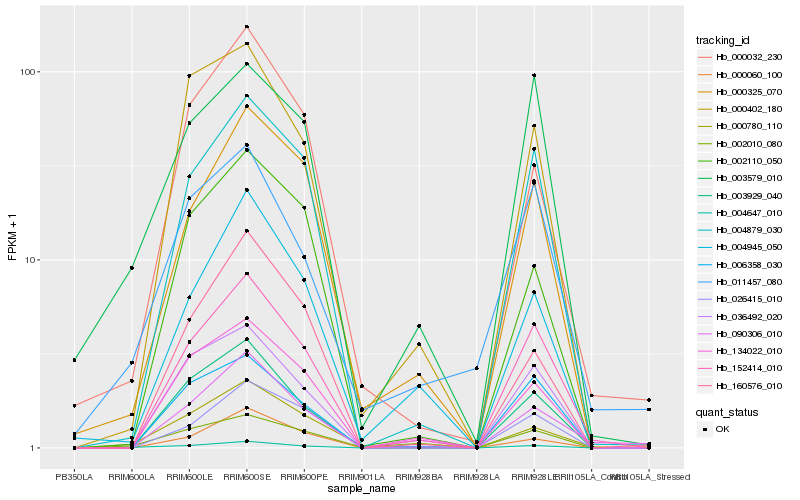
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004647_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000780_110 |
0.1556913463 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 3 |
Hb_003929_040 |
0.1630259306 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004879_030 |
0.1701296368 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 5 |
Hb_152414_010 |
0.1767591254 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 6 |
Hb_000032_230 |
0.1804141432 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |
| 7 |
Hb_003579_010 |
0.1805419624 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 8 |
Hb_000325_070 |
0.1854448341 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_002010_080 |
0.1895924613 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 10 |
Hb_006358_030 |
0.1913609125 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 11 |
Hb_036492_020 |
0.1917414656 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 12 |
Hb_000402_180 |
0.1928490291 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 13 |
Hb_002110_050 |
0.1928704641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_134022_010 |
0.1934760659 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 15 |
Hb_090306_010 |
0.194993257 |
- |
- |
hypothetical protein POPTR_0007s02300g [Populus trichocarpa] |
| 16 |
Hb_160576_010 |
0.1956568599 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 17 |
Hb_026415_010 |
0.1965152598 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011457_080 |
0.1973232858 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004945_050 |
0.200974384 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 20 |
Hb_000060_100 |
0.2031371605 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |