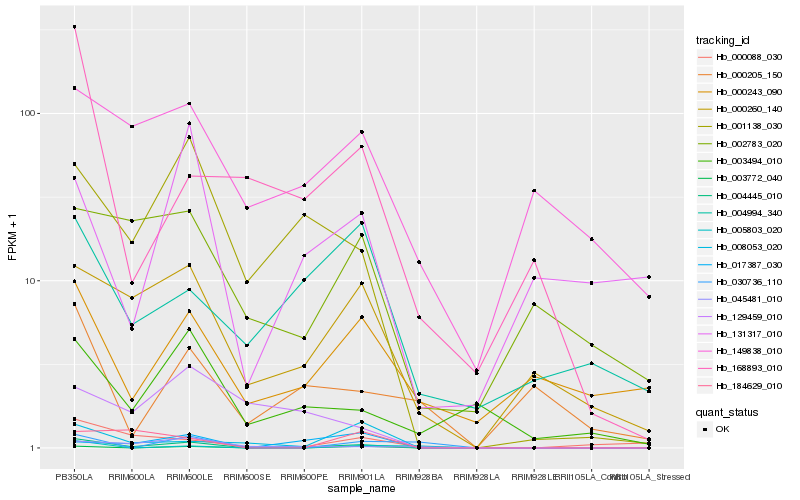
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004445_010 |
0.0 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 2 |
Hb_001138_030 |
0.2806345211 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 3 |
Hb_003772_040 |
0.2821400752 |
- |
- |
- |
| 4 |
Hb_000260_140 |
0.3069114818 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 5 |
Hb_003494_010 |
0.3373246864 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |
| 6 |
Hb_045481_010 |
0.3386070003 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_008053_020 |
0.346747837 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_168893_010 |
0.3489335438 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 9 |
Hb_000088_030 |
0.3500827929 |
- |
- |
hypothetical protein JCGZ_10947 [Jatropha curcas] |
| 10 |
Hb_017387_030 |
0.3527887567 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 11 |
Hb_000243_090 |
0.3576093961 |
- |
- |
PREDICTED: protein RTE1-HOMOLOG isoform X1 [Jatropha curcas] |
| 12 |
Hb_030736_110 |
0.3594542724 |
- |
- |
- |
| 13 |
Hb_184629_010 |
0.3595728484 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_149838_010 |
0.3630597453 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 15 |
Hb_004994_340 |
0.3671899463 |
- |
- |
- |
| 16 |
Hb_131317_010 |
0.3677778932 |
- |
- |
PREDICTED: PCTP-like protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_002783_020 |
0.3682658932 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X4 [Jatropha curcas] |
| 18 |
Hb_000205_150 |
0.3705767633 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 19 |
Hb_005803_020 |
0.3743144961 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 20 |
Hb_129459_010 |
0.3766833127 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |