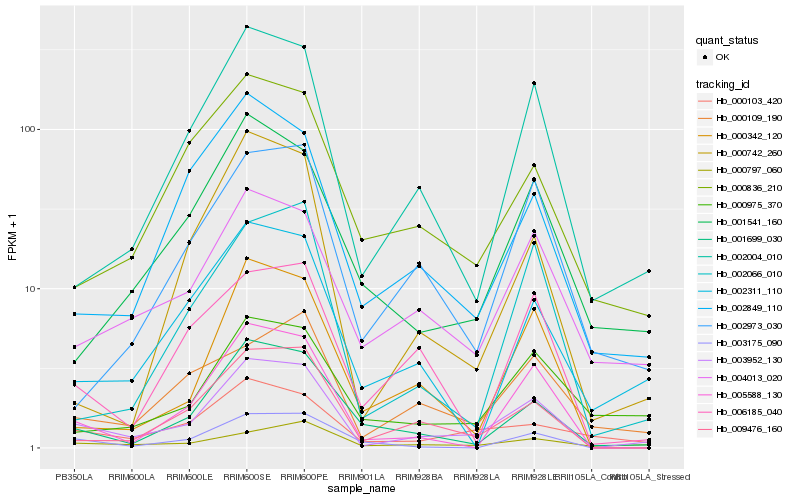
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003952_130 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 2 |
Hb_009476_160 |
0.169544441 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 3 |
Hb_002004_010 |
0.1706523772 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 4 |
Hb_005588_130 |
0.1743966529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001699_030 |
0.1768375446 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004013_020 |
0.1809110996 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 7 |
Hb_002311_110 |
0.1956773763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000342_120 |
0.1997526966 |
- |
- |
PREDICTED: probable protein phosphatase 2C 72 [Jatropha curcas] |
| 9 |
Hb_000103_420 |
0.1999313372 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Populus euphratica] |
| 10 |
Hb_002066_010 |
0.2014701802 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002849_110 |
0.2020277615 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 12 |
Hb_001541_160 |
0.2020825831 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 13 |
Hb_000975_370 |
0.2027309287 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000836_210 |
0.2029871048 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 15 |
Hb_003175_090 |
0.204882808 |
- |
- |
hypothetical protein POPTR_0011s01120g [Populus trichocarpa] |
| 16 |
Hb_000742_260 |
0.2056224269 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 17 |
Hb_000797_060 |
0.2061924756 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 18 |
Hb_000109_190 |
0.2066255833 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 19 |
Hb_002973_030 |
0.2082872722 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_006185_040 |
0.2089517382 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |