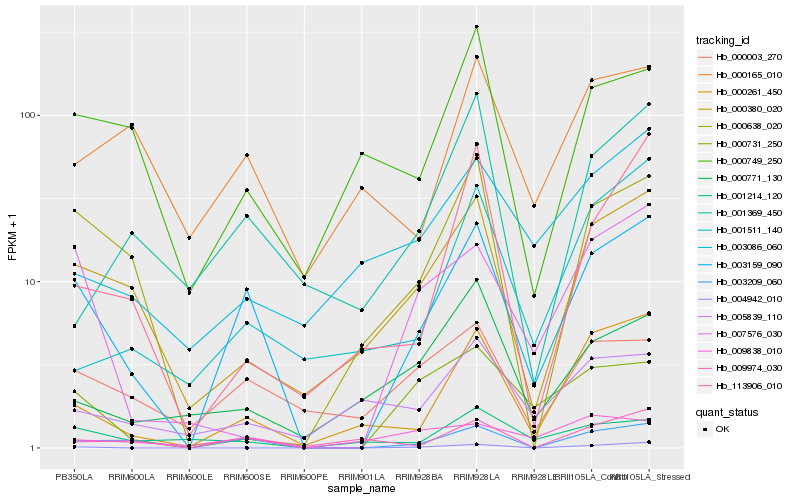
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003159_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_003209_060 |
0.155253762 |
- |
- |
- |
| 3 |
Hb_000261_450 |
0.2179919955 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000380_020 |
0.2281161298 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 5 |
Hb_000003_270 |
0.2466254823 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 6 |
Hb_000731_250 |
0.2479575032 |
- |
- |
- |
| 7 |
Hb_007576_030 |
0.2581332614 |
- |
- |
- |
| 8 |
Hb_001214_120 |
0.2641029313 |
- |
- |
- |
| 9 |
Hb_009974_030 |
0.2669750756 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_000165_010 |
0.2707260429 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 11 |
Hb_000749_250 |
0.2742680796 |
- |
- |
PREDICTED: lanC-like protein GCR2 [Jatropha curcas] |
| 12 |
Hb_001511_140 |
0.2751411824 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 13 |
Hb_113906_010 |
0.275682379 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 14 |
Hb_001369_450 |
0.2778721521 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_003086_060 |
0.2806428443 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 16 |
Hb_009838_010 |
0.2808072918 |
- |
- |
- |
| 17 |
Hb_000638_020 |
0.2810499638 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 18 |
Hb_004942_010 |
0.2814069328 |
- |
- |
hypothetical protein AMTR_s00057p00201180 [Amborella trichopoda] |
| 19 |
Hb_000771_130 |
0.2814311486 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_005839_110 |
0.2820069709 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Populus euphratica] |