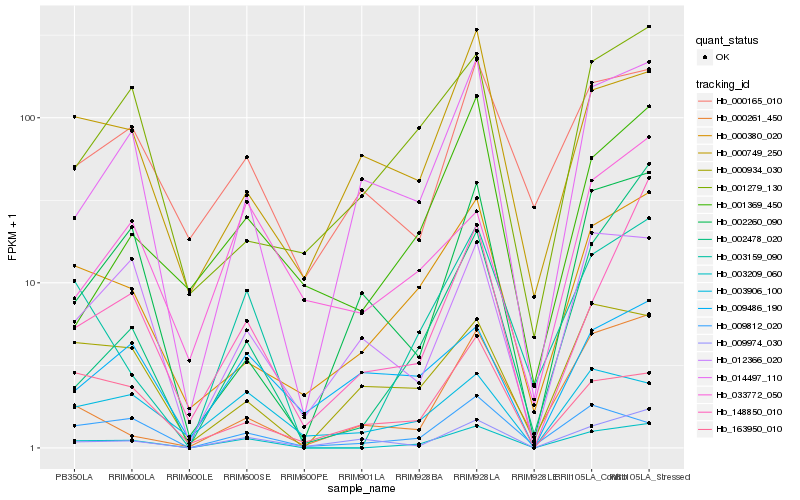
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003209_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_003159_090 |
0.155253762 |
- |
- |
- |
| 3 |
Hb_014497_110 |
0.2106920415 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 4 |
Hb_000380_020 |
0.2158058662 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 5 |
Hb_009974_030 |
0.2224208743 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_012366_020 |
0.2293894021 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003906_100 |
0.2308459313 |
- |
- |
hypothetical protein JCGZ_26174 [Jatropha curcas] |
| 8 |
Hb_148850_010 |
0.2349705276 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 9 |
Hb_009812_020 |
0.2382065238 |
- |
- |
- |
| 10 |
Hb_000934_030 |
0.2434952606 |
- |
- |
- |
| 11 |
Hb_000261_450 |
0.2450950045 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000165_010 |
0.2452360819 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 13 |
Hb_001369_450 |
0.2473705865 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_002260_090 |
0.248255723 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002478_020 |
0.2493748176 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 16 |
Hb_001279_130 |
0.250434976 |
- |
- |
hypothetical protein L484_017676 [Morus notabilis] |
| 17 |
Hb_033772_050 |
0.2528804632 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 18 |
Hb_009486_190 |
0.253638883 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_163950_010 |
0.2555572396 |
- |
- |
- |
| 20 |
Hb_000749_250 |
0.257455577 |
- |
- |
PREDICTED: lanC-like protein GCR2 [Jatropha curcas] |