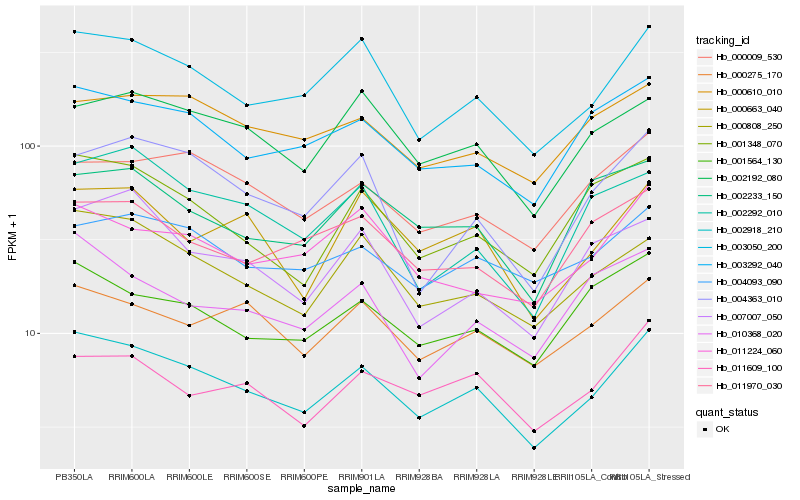
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |
| 2 |
Hb_003050_200 |
0.0649342595 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 3 |
Hb_000808_250 |
0.0800118471 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 4 |
Hb_003292_040 |
0.0870974843 |
transcription factor |
TF Family: MBF1 |
uncharacterized protein LOC100500420 [Glycine max] |
| 5 |
Hb_001348_070 |
0.0936673451 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_001564_130 |
0.0950363567 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_000663_040 |
0.0964328811 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000009_530 |
0.0972917168 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 9 |
Hb_004363_010 |
0.0974011111 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 10 |
Hb_000275_170 |
0.0979197739 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 11 |
Hb_002192_080 |
0.0987253709 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 12 |
Hb_004093_090 |
0.0988716409 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 13 |
Hb_011609_100 |
0.0999715306 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_011224_060 |
0.1000751681 |
- |
- |
hypothetical protein POPTR_0011s16880g [Populus trichocarpa] |
| 15 |
Hb_011970_030 |
0.1021340444 |
- |
- |
PREDICTED: uncharacterized protein LOC105641805 [Jatropha curcas] |
| 16 |
Hb_007007_050 |
0.104687884 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 17 |
Hb_000610_010 |
0.1047391626 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 18 |
Hb_002292_010 |
0.1048435446 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 19 |
Hb_010368_020 |
0.1052243146 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 20 |
Hb_002233_150 |
0.1056695421 |
- |
- |
PREDICTED: uncharacterized protein LOC105633093 [Jatropha curcas] |