| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
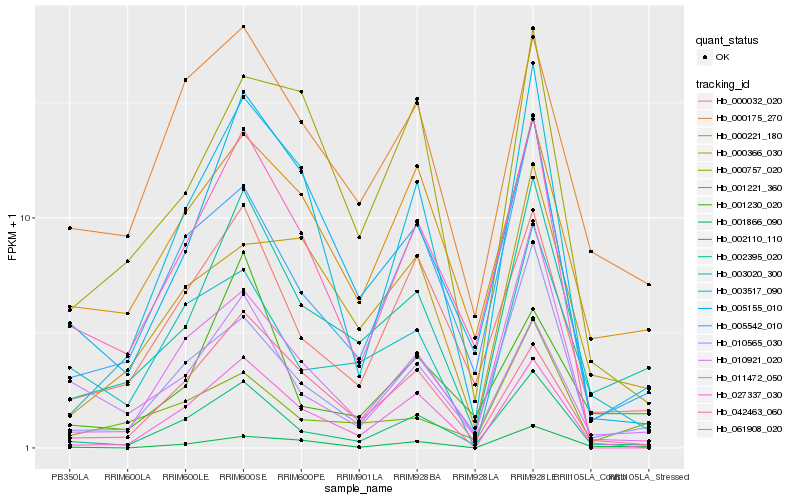
Hb_002395_020 |
0.0 |
- |
- |
Protein CREG1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002110_110 |
0.1375342943 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 3 |
Hb_000032_020 |
0.166146049 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 4 |
Hb_000221_180 |
0.1682764849 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001221_360 |
0.1723563692 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 6 |
Hb_010565_030 |
0.1747673662 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 7 |
Hb_000175_270 |
0.175848697 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005155_010 |
0.1781827649 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_005542_010 |
0.1797953369 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 10 |
Hb_003517_090 |
0.1798222441 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001230_020 |
0.1875287362 |
- |
- |
hypothetical protein POPTR_0001s31610g [Populus trichocarpa] |
| 12 |
Hb_003020_300 |
0.1880955582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_027337_030 |
0.1901437368 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 14 |
Hb_000366_030 |
0.1908000068 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 15 |
Hb_061908_020 |
0.1938418014 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 16 |
Hb_001866_090 |
0.1940344272 |
- |
- |
hypothetical protein VITISV_023491 [Vitis vinifera] |
| 17 |
Hb_011472_050 |
0.1949688098 |
- |
- |
- |
| 18 |
Hb_010921_020 |
0.1960369032 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 19 |
Hb_000757_020 |
0.1960596266 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_042463_060 |
0.1964330423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |