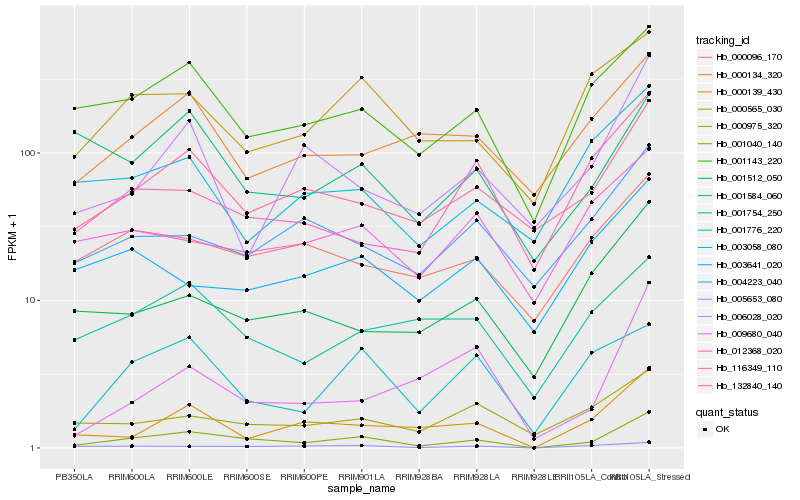
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_132840_140 |
0.2037446709 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1 [Vitis vinifera] |
| 3 |
Hb_001143_220 |
0.2041402016 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 4 |
Hb_000139_430 |
0.2081315279 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 5 |
Hb_005653_080 |
0.2126406031 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_001776_220 |
0.2272158909 |
- |
- |
- |
| 7 |
Hb_001512_050 |
0.22764394 |
- |
- |
copine, putative [Ricinus communis] |
| 8 |
Hb_000565_030 |
0.2292129471 |
- |
- |
PREDICTED: 40S ribosomal protein S26-3-like [Jatropha curcas] |
| 9 |
Hb_006028_020 |
0.2307187201 |
- |
- |
PREDICTED: 60S ribosomal protein L38-like [Cicer arietinum] |
| 10 |
Hb_001754_250 |
0.233261683 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 11 |
Hb_000096_170 |
0.2345125554 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Jatropha curcas] |
| 12 |
Hb_000975_320 |
0.2352890501 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004223_040 |
0.2361532202 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 14 |
Hb_000134_320 |
0.2380876015 |
- |
- |
hypothetical protein PRUPE_ppa013450mg [Prunus persica] |
| 15 |
Hb_116349_110 |
0.2388839735 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 16 |
Hb_012368_020 |
0.2420821092 |
- |
- |
PREDICTED: uncharacterized protein LOC105650964 [Jatropha curcas] |
| 17 |
Hb_009680_040 |
0.2423319352 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001584_060 |
0.2440983624 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 19 |
Hb_003058_080 |
0.2442101338 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 20 |
Hb_003641_020 |
0.2462670761 |
- |
- |
hypothetical protein POPTR_0006s13960g [Populus trichocarpa] |