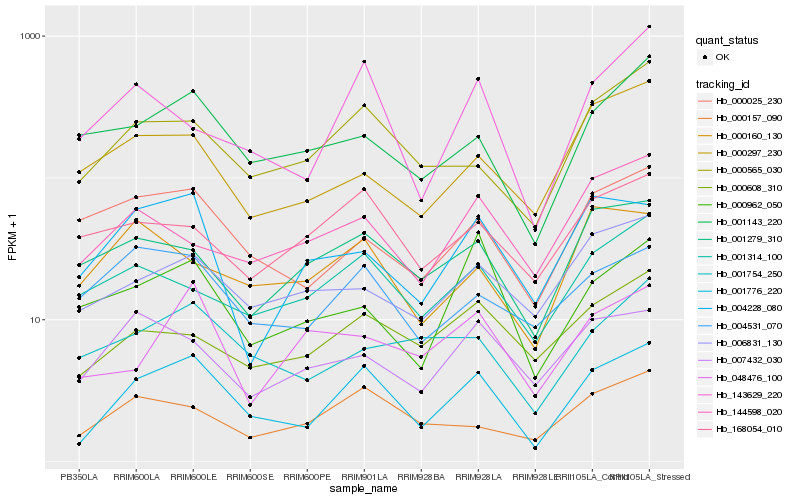
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_000565_030 |
0.1765822427 |
- |
- |
PREDICTED: 40S ribosomal protein S26-3-like [Jatropha curcas] |
| 3 |
Hb_000157_090 |
0.188884086 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 4 |
Hb_143629_220 |
0.1903232355 |
- |
- |
PREDICTED: hydrophobic protein LTI6B [Phoenix dactylifera] |
| 5 |
Hb_000608_310 |
0.1923665416 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_001143_220 |
0.1935830988 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 7 |
Hb_000025_230 |
0.1944332132 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |
| 8 |
Hb_004531_070 |
0.1953061791 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX19-like [Jatropha curcas] |
| 9 |
Hb_006831_130 |
0.1957153919 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_001754_250 |
0.1959491961 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 11 |
Hb_001314_100 |
0.197779824 |
- |
- |
hypothetical protein POPTR_0001s01820g [Populus trichocarpa] |
| 12 |
Hb_004228_080 |
0.2003863878 |
- |
- |
phosphoethanolamine n-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001279_310 |
0.2014669191 |
- |
- |
hypothetical protein JCGZ_10212 [Jatropha curcas] |
| 14 |
Hb_007432_030 |
0.2020694802 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 15 |
Hb_000962_050 |
0.2050796553 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 16 |
Hb_048476_100 |
0.2072293654 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |
| 17 |
Hb_000297_230 |
0.2079890211 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 18 |
Hb_168054_010 |
0.2085105133 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP18-1 [Elaeis guineensis] |
| 19 |
Hb_144598_020 |
0.2096549791 |
- |
- |
PREDICTED: triphosphate tunel metalloenzyme 3 [Jatropha curcas] |
| 20 |
Hb_000160_130 |
0.2100991489 |
- |
- |
PREDICTED: uncharacterized protein LOC105648215 [Jatropha curcas] |