| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
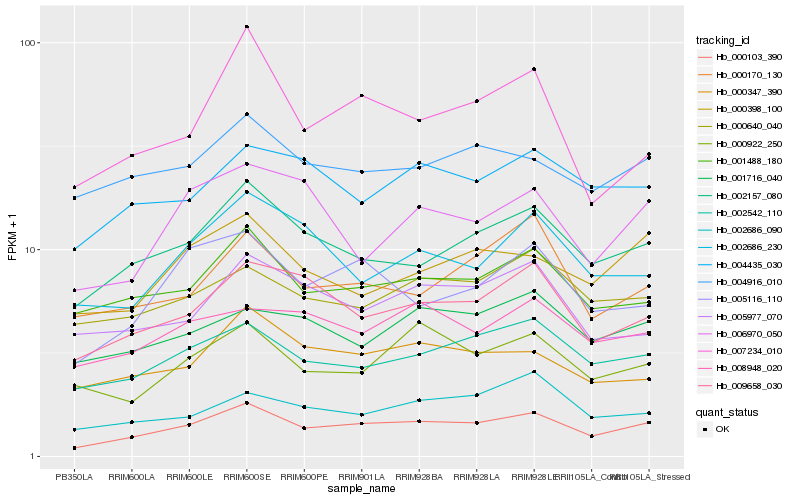
Hb_000103_390 |
0.0 |
- |
- |
Pentatricopeptide repeat superfamily protein [Theobroma cacao] |
| 2 |
Hb_002157_080 |
0.0915179015 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002542_110 |
0.095074545 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 4 |
Hb_009658_030 |
0.0997621879 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 5 |
Hb_001488_180 |
0.1032226149 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005116_110 |
0.1047062681 |
- |
- |
PREDICTED: uncharacterized protein LOC105640215 [Jatropha curcas] |
| 7 |
Hb_004435_030 |
0.111319941 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 8 |
Hb_000347_390 |
0.1122291728 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 9 |
Hb_000398_100 |
0.1135385275 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 10 |
Hb_000922_250 |
0.1143127552 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_008948_020 |
0.1153976498 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 12 |
Hb_000640_040 |
0.1164747188 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004916_010 |
0.1168226238 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 14 |
Hb_001716_040 |
0.1175752253 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 15 |
Hb_002686_090 |
0.1177666464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 16 |
Hb_000170_130 |
0.1187109297 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 17 |
Hb_007234_010 |
0.119665781 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |
| 18 |
Hb_002686_230 |
0.1203151247 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_006970_050 |
0.1207365331 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 20 |
Hb_005977_070 |
0.1210926625 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |