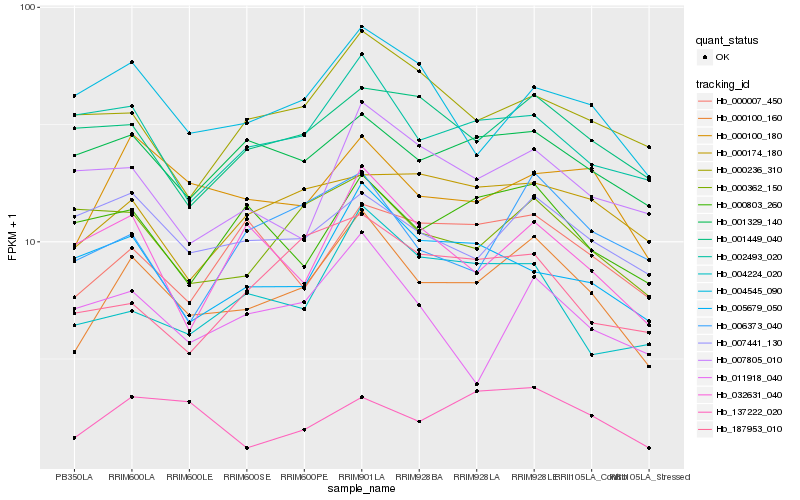
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000100_160 |
0.0 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 2 |
Hb_000100_180 |
0.116197382 |
- |
- |
PREDICTED: probable acyl-activating enzyme 16, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004545_090 |
0.1377488354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002493_020 |
0.1393736732 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000007_450 |
0.1395465142 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001329_140 |
0.1413540515 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 7 |
Hb_004224_020 |
0.1421255889 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 8 |
Hb_000362_150 |
0.1428235396 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 9 |
Hb_001449_040 |
0.1436195993 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000174_180 |
0.144066258 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_006373_040 |
0.1442660138 |
- |
- |
PREDICTED: potassium transporter 11-like [Nicotiana tomentosiformis] |
| 12 |
Hb_005679_050 |
0.1448978838 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000236_310 |
0.1452477073 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 14 |
Hb_000803_260 |
0.1457525598 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 15 |
Hb_032631_040 |
0.1463448762 |
- |
- |
PREDICTED: uncharacterized protein LOC105638769 [Jatropha curcas] |
| 16 |
Hb_007441_130 |
0.147022868 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_011918_040 |
0.1471484937 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 18 |
Hb_137222_020 |
0.1494553925 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 19 |
Hb_007805_010 |
0.1494764867 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 20 |
Hb_187953_010 |
0.1495970021 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |