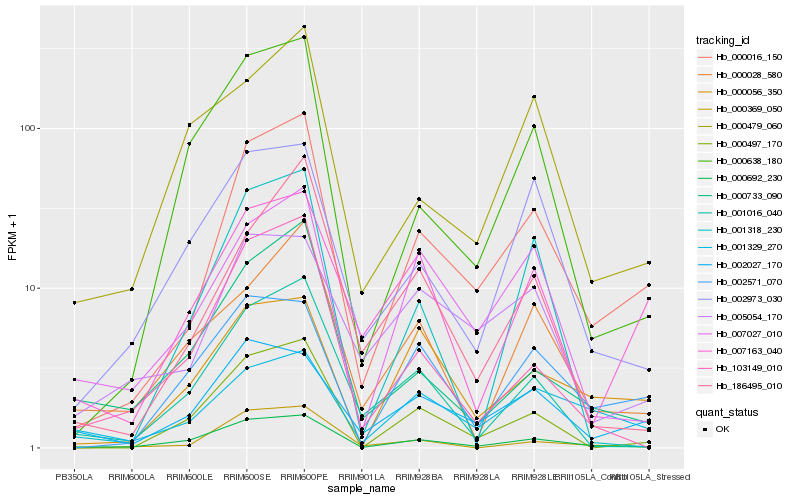
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105137629 [Populus euphratica] |
| 2 |
Hb_000692_230 |
0.1664182812 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 3 |
Hb_000497_170 |
0.1770727077 |
- |
- |
PREDICTED: uncharacterized protein LOC105647275 [Jatropha curcas] |
| 4 |
Hb_000369_050 |
0.1933751188 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 5 |
Hb_000638_180 |
0.1979709413 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 6 |
Hb_007027_010 |
0.1995872397 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 7 |
Hb_186495_010 |
0.2014281506 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 8 |
Hb_000056_350 |
0.2023601172 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 9 |
Hb_002571_070 |
0.2026871991 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 10 |
Hb_002027_170 |
0.202846111 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 11 |
Hb_007163_040 |
0.2054786773 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 12 |
Hb_005054_170 |
0.2112821301 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001016_040 |
0.2113619649 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002973_030 |
0.2125407237 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_103149_010 |
0.2163313472 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 16 |
Hb_001318_230 |
0.2166593684 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 17 |
Hb_000733_090 |
0.2173998596 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 18 |
Hb_001329_270 |
0.2197883848 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 19 |
Hb_000028_580 |
0.2198848761 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 20 |
Hb_000479_060 |
0.2200591227 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |