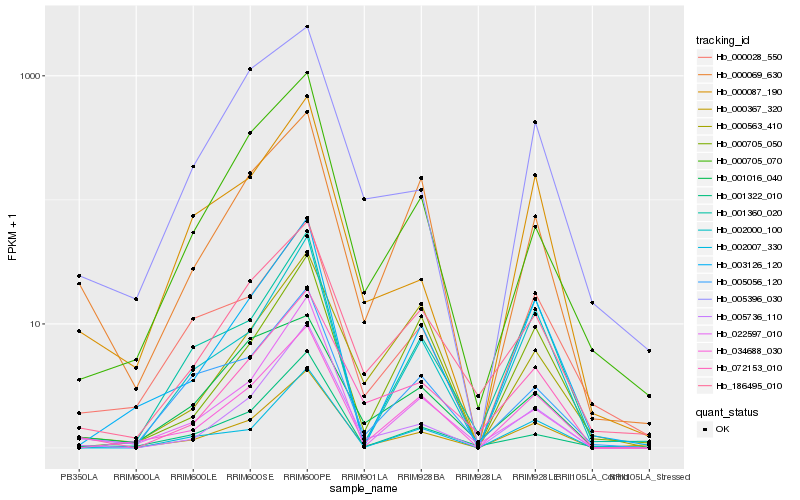
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_072153_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 2 |
Hb_186495_010 |
0.1023884948 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 3 |
Hb_005736_110 |
0.1439859321 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 4 |
Hb_000367_320 |
0.1588959325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_034688_030 |
0.1633172785 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 6 |
Hb_000069_630 |
0.179011999 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 7 |
Hb_000563_410 |
0.1796388925 |
- |
- |
hypothetical protein JCGZ_18917 [Jatropha curcas] |
| 8 |
Hb_003126_120 |
0.187359699 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 9 |
Hb_005396_030 |
0.190597374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005056_120 |
0.1913660571 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_002000_100 |
0.1923697883 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 12 |
Hb_001016_040 |
0.1932349945 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002007_330 |
0.193372431 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 5 [Jatropha curcas] |
| 14 |
Hb_022597_010 |
0.1935812714 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 15 |
Hb_000028_550 |
0.1953568586 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_001360_020 |
0.1959206359 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 17 |
Hb_000705_070 |
0.1961696794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000087_190 |
0.1968791171 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 19 |
Hb_000705_050 |
0.1980964177 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 20 |
Hb_001322_010 |
0.1993795012 |
- |
- |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |