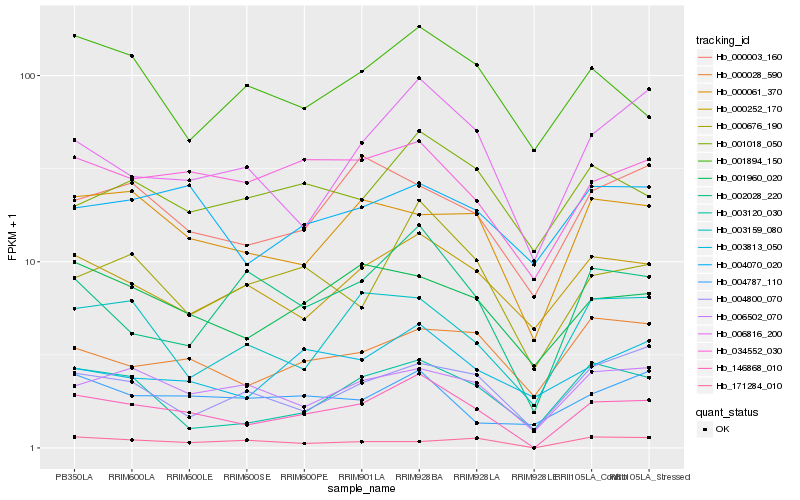
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146868_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_034552_030 |
0.1607025896 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_000676_190 |
0.1636828801 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |
| 4 |
Hb_001960_020 |
0.1641298727 |
- |
- |
PREDICTED: mitochondrial arginine transporter BAC2 [Jatropha curcas] |
| 5 |
Hb_006502_070 |
0.1645314353 |
- |
- |
separase, putative [Ricinus communis] |
| 6 |
Hb_000061_370 |
0.1655169218 |
- |
- |
PREDICTED: zinc finger RNA-binding protein [Vitis vinifera] |
| 7 |
Hb_002028_220 |
0.1684859174 |
- |
- |
aminoadipic semialdehyde synthase, putative [Ricinus communis] |
| 8 |
Hb_000252_170 |
0.1731114883 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006816_200 |
0.1732892255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003120_030 |
0.1760344417 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 11 |
Hb_003159_080 |
0.1769440534 |
- |
- |
PREDICTED: uncharacterized protein LOC105639276 [Jatropha curcas] |
| 12 |
Hb_000003_160 |
0.1819682936 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 13 |
Hb_003813_050 |
0.1824453539 |
- |
- |
PREDICTED: tRNA-specific adenosine deaminase 2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000028_590 |
0.1847470503 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 15 |
Hb_004800_070 |
0.1847661766 |
- |
- |
- |
| 16 |
Hb_001018_050 |
0.1857498214 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 17 |
Hb_004787_110 |
0.1859451401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004070_020 |
0.1863747749 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 19 |
Hb_171284_010 |
0.1867471457 |
- |
- |
- |
| 20 |
Hb_001894_150 |
0.1887631251 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |