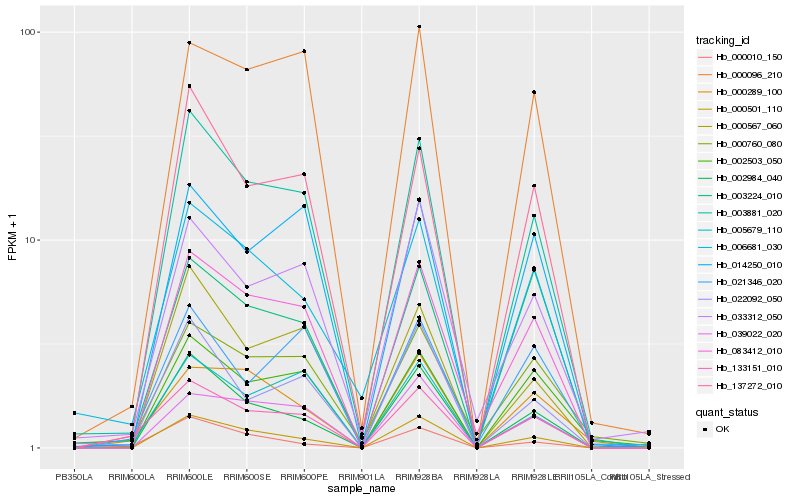
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133151_010 |
0.0 |
- |
- |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |
| 2 |
Hb_003881_020 |
0.1363620451 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 3 |
Hb_083412_010 |
0.1553748752 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 4 |
Hb_000289_100 |
0.1569501315 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 5 |
Hb_005679_110 |
0.1583227083 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 6 |
Hb_137272_010 |
0.1613883755 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 7 |
Hb_000501_110 |
0.1617404542 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 8 |
Hb_006681_030 |
0.1654695491 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000010_150 |
0.1656578684 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Jatropha curcas] |
| 10 |
Hb_002984_040 |
0.165735046 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_033312_050 |
0.169008665 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 12 |
Hb_022092_050 |
0.1708012221 |
- |
- |
PREDICTED: uncharacterized protein LOC105136214 [Populus euphratica] |
| 13 |
Hb_000096_210 |
0.1721245203 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 14 |
Hb_000760_080 |
0.1728091765 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 15 |
Hb_039022_020 |
0.173399681 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 16 |
Hb_002503_050 |
0.1736922018 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 17 |
Hb_021346_020 |
0.1754569728 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 18 |
Hb_003224_010 |
0.1755670703 |
- |
- |
- |
| 19 |
Hb_014250_010 |
0.1758090088 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000567_060 |
0.175912235 |
- |
- |
PREDICTED: U-box domain-containing protein 7 [Jatropha curcas] |