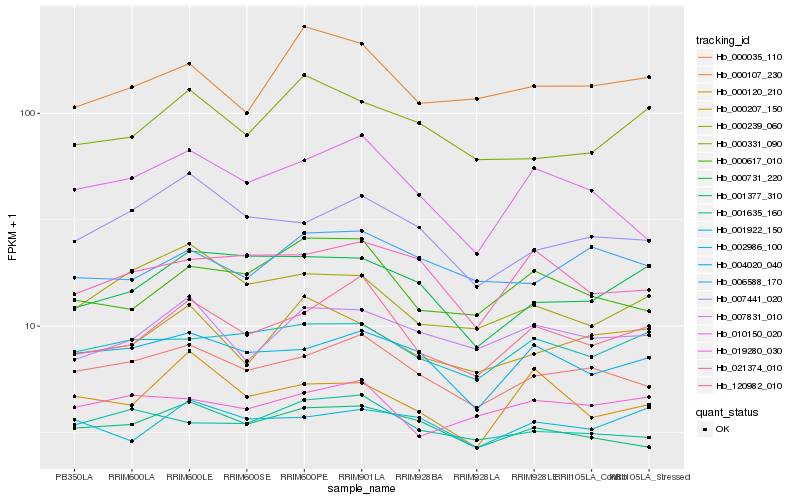
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_120982_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003116mg, partial [Citrus clementina] |
| 2 |
Hb_000035_110 |
0.0781657486 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 3 |
Hb_000617_010 |
0.0815282072 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 4 |
Hb_007831_010 |
0.0894133648 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 5 |
Hb_004020_040 |
0.0895466128 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001922_150 |
0.0901058185 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_010150_020 |
0.0929476051 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 8 |
Hb_000731_220 |
0.0938131143 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |
| 9 |
Hb_000107_230 |
0.094709781 |
- |
- |
unknown [Medicago truncatula] |
| 10 |
Hb_019280_030 |
0.0964655404 |
- |
- |
- |
| 11 |
Hb_001377_310 |
0.0966963957 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 12 |
Hb_021374_010 |
0.0967745869 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 13 |
Hb_002986_100 |
0.0991610944 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 14 |
Hb_007441_020 |
0.1003521349 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 15 |
Hb_006588_170 |
0.1017315308 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 16 |
Hb_000207_150 |
0.1029041377 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 17 |
Hb_000120_210 |
0.1031009728 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 18 |
Hb_001635_160 |
0.1037865203 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 19 |
Hb_000239_060 |
0.1050067709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000331_090 |
0.1053509689 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |