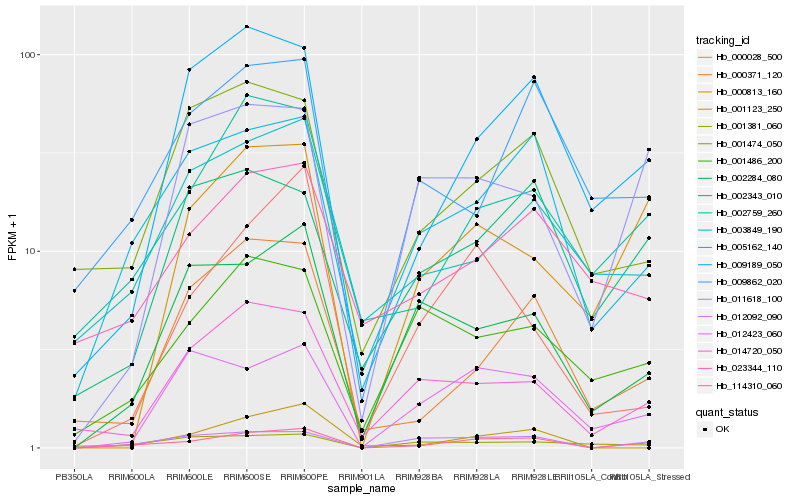
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114310_060 |
0.0 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2G, putative [Ricinus communis] |
| 2 |
Hb_012092_090 |
0.1771252711 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005162_140 |
0.1859654719 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 4 |
Hb_002759_260 |
0.2106507192 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 5 |
Hb_009189_050 |
0.2167073394 |
- |
- |
PREDICTED: uncharacterized protein LOC105648080 [Jatropha curcas] |
| 6 |
Hb_012423_060 |
0.2319600895 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 7 |
Hb_002284_080 |
0.2352082715 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_014720_050 |
0.2353982159 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 9 |
Hb_000371_120 |
0.2378100322 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_000813_160 |
0.2378955983 |
- |
- |
hypothetical protein PRUPE_ppa013181mg [Prunus persica] |
| 11 |
Hb_011618_100 |
0.2417280055 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 12 |
Hb_001474_050 |
0.2426171272 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 13 |
Hb_009862_020 |
0.2453285891 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 14 |
Hb_001123_250 |
0.2461989908 |
- |
- |
hypothetical protein POPTR_0001s13220g [Populus trichocarpa] |
| 15 |
Hb_002343_010 |
0.2466129543 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_003849_190 |
0.2489562524 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 17 |
Hb_000028_500 |
0.2502828255 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_001486_200 |
0.2509090601 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 19 |
Hb_001381_060 |
0.2512270361 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 20 |
Hb_023344_110 |
0.2517885717 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |