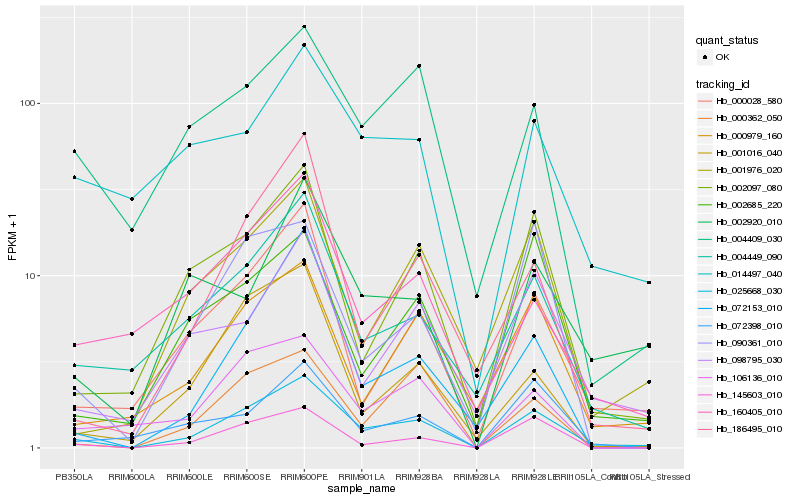
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025668_030 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 2 |
Hb_004449_090 |
0.1779556404 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 3 |
Hb_145603_010 |
0.1780075702 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_000028_580 |
0.1820189848 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 5 |
Hb_002097_080 |
0.1848267247 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 6 |
Hb_000979_160 |
0.1911162214 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 7 |
Hb_001976_020 |
0.1945608247 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_106136_010 |
0.1953943902 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_098795_030 |
0.1978706795 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 10 |
Hb_072398_010 |
0.2039492428 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 11 |
Hb_004409_030 |
0.2044116859 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |
| 12 |
Hb_000362_050 |
0.205462697 |
transcription factor |
TF Family: NOZZLE |
PREDICTED: uncharacterized protein LOC105635737 [Jatropha curcas] |
| 13 |
Hb_002685_220 |
0.207207388 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_186495_010 |
0.2076287976 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 15 |
Hb_072153_010 |
0.2093926157 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 16 |
Hb_090361_010 |
0.2102123394 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_160405_010 |
0.2114415557 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_002920_010 |
0.2122726503 |
- |
- |
PREDICTED: sulfiredoxin, chloroplastic/mitochondrial [Populus euphratica] |
| 19 |
Hb_014497_040 |
0.2148828461 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001016_040 |
0.2154824407 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |