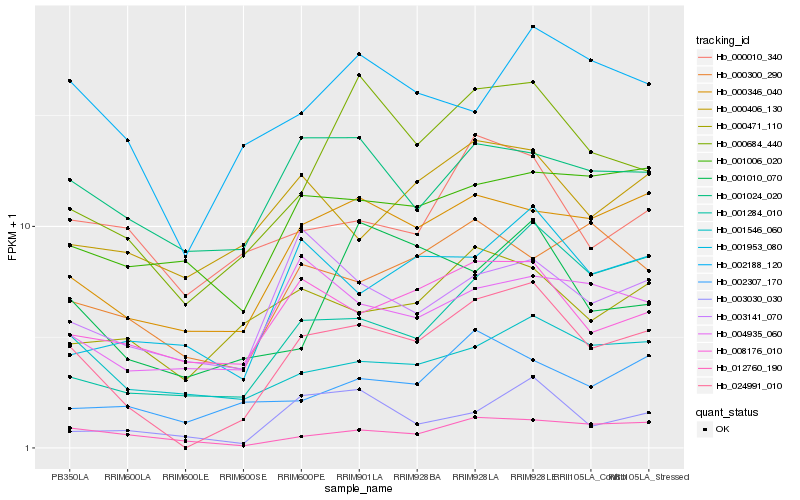
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024991_010 |
0.0 |
- |
- |
hypothetical protein B456_007G217100 [Gossypium raimondii] |
| 2 |
Hb_000684_440 |
0.1700024078 |
- |
- |
hypothetical protein CICLE_v100111082mg, partial [Citrus clementina] |
| 3 |
Hb_000346_040 |
0.1746613428 |
- |
- |
- |
| 4 |
Hb_008176_010 |
0.1931592167 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 5 |
Hb_012760_190 |
0.1971161732 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 6 |
Hb_002188_120 |
0.1976372765 |
- |
- |
PREDICTED: thymocyte nuclear protein 1 [Pyrus x bretschneideri] |
| 7 |
Hb_001953_080 |
0.2009795478 |
- |
- |
pseudouridine synthase family protein, partial [Manihot esculenta] |
| 8 |
Hb_001284_010 |
0.2021425924 |
- |
- |
hypothetical protein CISIN_1g0279981mg, partial [Citrus sinensis] |
| 9 |
Hb_000471_110 |
0.2044832126 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004935_060 |
0.2073702749 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_003030_030 |
0.2077976276 |
- |
- |
- |
| 12 |
Hb_001546_060 |
0.2087428216 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 13 |
Hb_000406_130 |
0.2092085035 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 14 |
Hb_003141_070 |
0.2096606398 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 15 |
Hb_001010_070 |
0.2105233492 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 16 |
Hb_001006_020 |
0.2105818396 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 17 |
Hb_000300_290 |
0.2108461647 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 18 |
Hb_001024_020 |
0.2114153365 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 19 |
Hb_002307_170 |
0.2119992919 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_000010_340 |
0.2126713576 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |