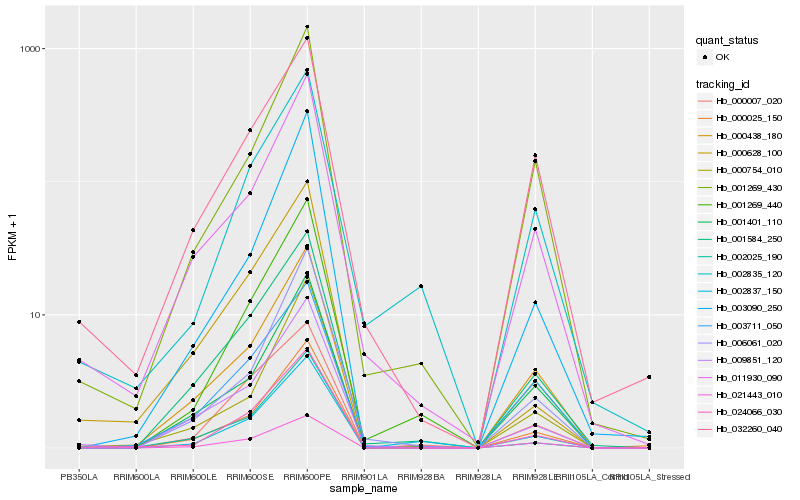
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024066_030 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 2 |
Hb_001269_440 |
0.1034129583 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 3 |
Hb_000438_180 |
0.1063061776 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000025_150 |
0.1090801052 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 5 |
Hb_021443_010 |
0.1107622043 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 6 |
Hb_001584_250 |
0.1113106024 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 7 |
Hb_002025_190 |
0.1127873457 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 8 |
Hb_000007_020 |
0.1128204472 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 9 |
Hb_001269_430 |
0.1169797724 |
- |
- |
hypothetical protein JCGZ_04286 [Jatropha curcas] |
| 10 |
Hb_011930_090 |
0.1173089599 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 11 |
Hb_009851_120 |
0.1194654204 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 12 |
Hb_000628_100 |
0.1214508789 |
- |
- |
hypothetical protein MTR_1g019660 [Medicago truncatula] |
| 13 |
Hb_003711_050 |
0.1238217064 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 14 |
Hb_000754_010 |
0.1250004324 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_002835_120 |
0.1259019666 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 16 |
Hb_032260_040 |
0.1259362195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002837_150 |
0.1264640865 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 18 |
Hb_003090_250 |
0.1268464767 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_001401_110 |
0.1293312257 |
- |
- |
hypothetical protein JCGZ_04272 [Jatropha curcas] |
| 20 |
Hb_006061_020 |
0.131888996 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |