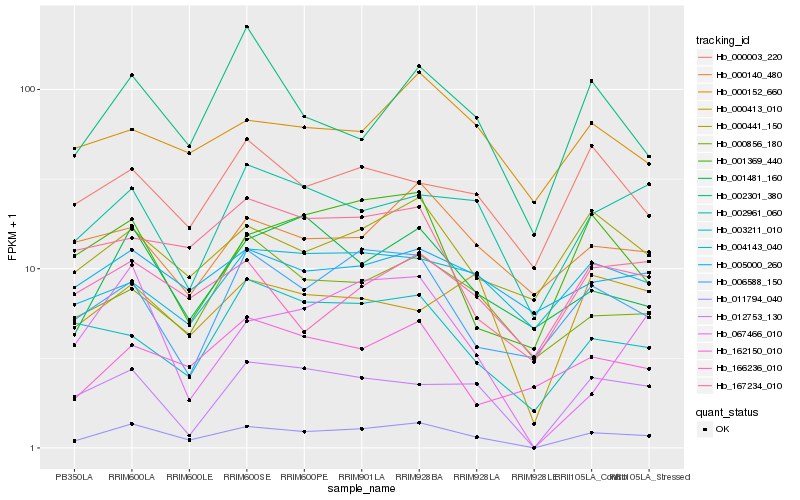
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011794_040 |
0.0 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 2 |
Hb_012753_130 |
0.1536917054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002961_060 |
0.1715443511 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 5 [Populus euphratica] |
| 4 |
Hb_000441_150 |
0.1734472741 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_167234_010 |
0.174252787 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 6 |
Hb_006588_150 |
0.1744453886 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 7 |
Hb_001369_440 |
0.1770040535 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 8 |
Hb_000856_180 |
0.1778290276 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 9 |
Hb_001481_160 |
0.1787178128 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 10 |
Hb_003211_010 |
0.1803465664 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 11 |
Hb_000413_010 |
0.1841937477 |
- |
- |
hypothetical protein JCGZ_21316 [Jatropha curcas] |
| 12 |
Hb_000140_480 |
0.1843984937 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 13 |
Hb_002301_380 |
0.185427636 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 14 |
Hb_067466_010 |
0.1856212384 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 15 |
Hb_000003_220 |
0.1871315389 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 16 |
Hb_000152_660 |
0.1888794967 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 17 |
Hb_166236_010 |
0.1902364398 |
- |
- |
- |
| 18 |
Hb_005000_260 |
0.1903198512 |
- |
- |
recA family protein [Populus trichocarpa] |
| 19 |
Hb_004143_040 |
0.1910592188 |
- |
- |
argininosuccinate lyase, putative [Ricinus communis] |
| 20 |
Hb_162150_010 |
0.191447194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |