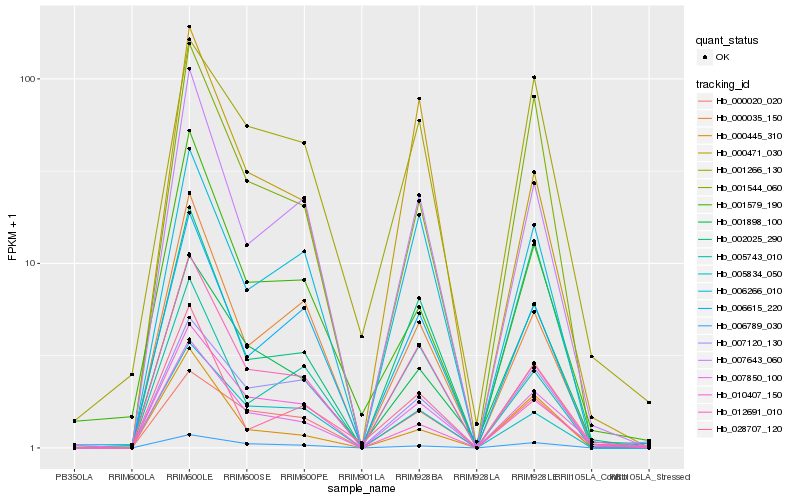
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007850_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 2 |
Hb_012691_010 |
0.1314412011 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 3 |
Hb_001898_100 |
0.1374408581 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 4 |
Hb_007643_060 |
0.1393459778 |
- |
- |
- |
| 5 |
Hb_006266_010 |
0.1485119573 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_001266_130 |
0.1508702668 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 7 |
Hb_006789_030 |
0.1520742812 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 8 |
Hb_002025_290 |
0.1526442429 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000020_020 |
0.1530256548 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 10 |
Hb_001544_060 |
0.154201428 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006615_220 |
0.1544075127 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005834_050 |
0.1629688229 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 13 |
Hb_007120_130 |
0.1638340037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_028707_120 |
0.1649200448 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000471_030 |
0.1681459662 |
- |
- |
- |
| 16 |
Hb_001579_190 |
0.1686538836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000035_150 |
0.1695924639 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 18 |
Hb_010407_150 |
0.1698296718 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 19 |
Hb_000445_310 |
0.17315424 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 20 |
Hb_005743_010 |
0.1739177602 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |