| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
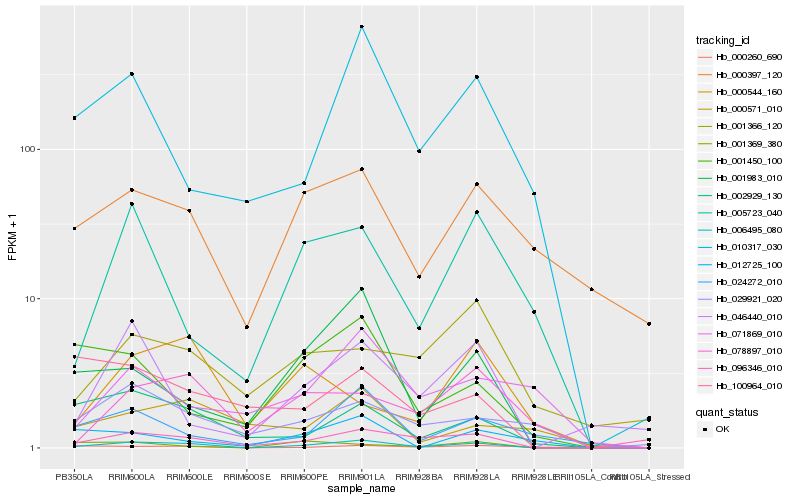
Hb_006495_080 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppb025438mg, partial [Prunus persica] |
| 2 |
Hb_002929_130 |
0.2725299262 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 3 |
Hb_096346_010 |
0.2737597692 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_001983_010 |
0.2751572833 |
- |
- |
- |
| 5 |
Hb_000397_120 |
0.2798012117 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 6 |
Hb_024272_010 |
0.280860178 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 7 |
Hb_010317_030 |
0.2876814492 |
- |
- |
unknown [Picea sitchensis] |
| 8 |
Hb_046440_010 |
0.2894170339 |
- |
- |
hypothetical protein JCGZ_06190 [Jatropha curcas] |
| 9 |
Hb_078897_010 |
0.2929418169 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 10 |
Hb_029921_020 |
0.2967183533 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 11 |
Hb_005723_040 |
0.2982473259 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001450_100 |
0.3098194186 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 13 |
Hb_000260_690 |
0.315244901 |
- |
- |
disease resistance protein (CC-NBS-LRR class) family protein [Medicago truncatula] |
| 14 |
Hb_001369_380 |
0.317593514 |
- |
- |
F-box family protein, putative [Theobroma cacao] |
| 15 |
Hb_001366_120 |
0.318412212 |
- |
- |
unknown [Zea mays] |
| 16 |
Hb_100964_010 |
0.3206716706 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000571_010 |
0.3213833474 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Gossypium raimondii] |
| 18 |
Hb_071869_010 |
0.3257907298 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012725_100 |
0.3263303438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000544_160 |
0.3307694444 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |