| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
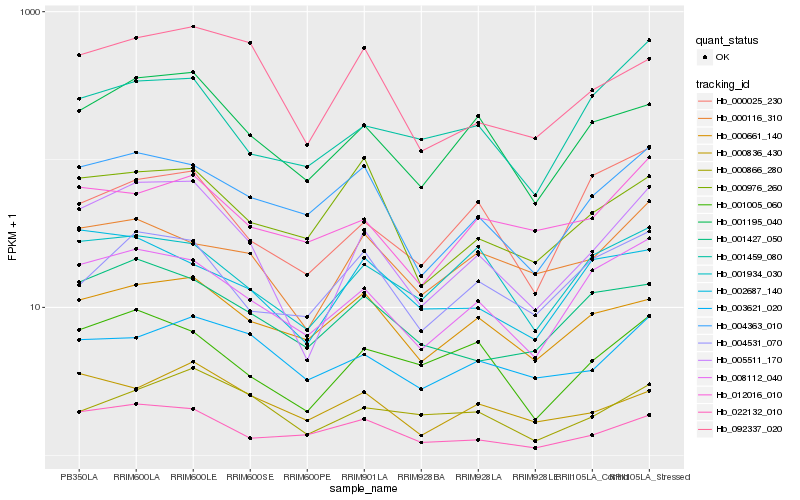
Hb_005511_170 |
0.0 |
- |
- |
hypothetical protein CISIN_1g030718mg [Citrus sinensis] |
| 2 |
Hb_001195_040 |
0.1250795587 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 3 |
Hb_008112_040 |
0.1279933041 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 4 |
Hb_001005_060 |
0.14055103 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 5 |
Hb_022132_010 |
0.1507679788 |
- |
- |
unknown [Medicago truncatula] |
| 6 |
Hb_004363_010 |
0.1509877616 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 7 |
Hb_092337_020 |
0.1526190124 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 8 |
Hb_000866_280 |
0.1560253977 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 9 |
Hb_001427_050 |
0.1579863689 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 10 |
Hb_001934_030 |
0.1597022238 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 11 |
Hb_000116_310 |
0.160770127 |
- |
- |
50S robosomal protein L11, putative [Ricinus communis] |
| 12 |
Hb_004531_070 |
0.1644055158 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX19-like [Jatropha curcas] |
| 13 |
Hb_000661_140 |
0.1656783569 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003621_020 |
0.1657178419 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 15 |
Hb_000836_430 |
0.1660626559 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000025_230 |
0.1693283478 |
- |
- |
PREDICTED: 10 kDa chaperonin [Vitis vinifera] |
| 17 |
Hb_002687_140 |
0.1698465714 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 18 |
Hb_012016_010 |
0.1701346584 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000976_260 |
0.1704367771 |
- |
- |
- |
| 20 |
Hb_001459_080 |
0.1713027066 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |