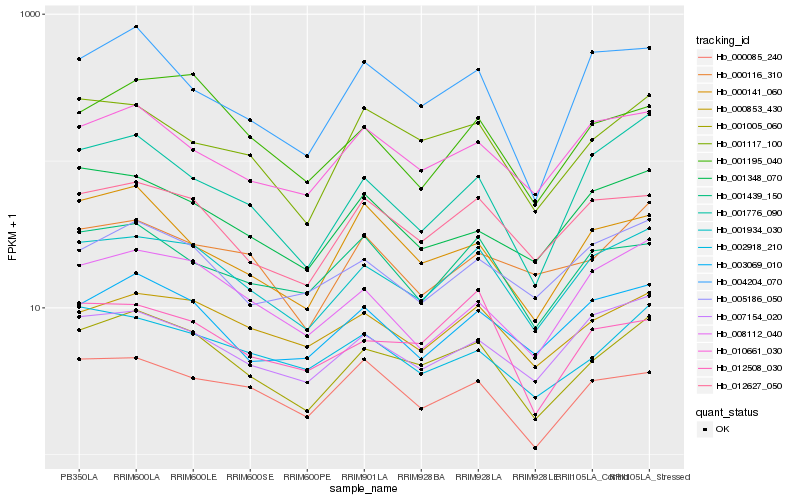
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001005_060 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 2 |
Hb_001934_030 |
0.0843356181 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 3 |
Hb_001117_100 |
0.107434864 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 4 |
Hb_004204_070 |
0.1176224939 |
- |
- |
PREDICTED: 40S ribosomal protein S19-3 [Jatropha curcas] |
| 5 |
Hb_012627_050 |
0.1177097923 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002918_210 |
0.1209283699 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |
| 7 |
Hb_003069_010 |
0.1229804627 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |
| 8 |
Hb_000853_430 |
0.1241090972 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 9 |
Hb_001776_090 |
0.1242415061 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 10 |
Hb_008112_040 |
0.125720074 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 11 |
Hb_007154_020 |
0.125936905 |
- |
- |
- |
| 12 |
Hb_000141_060 |
0.1261676157 |
- |
- |
PREDICTED: uncharacterized protein LOC105632248 [Jatropha curcas] |
| 13 |
Hb_010661_030 |
0.1300795961 |
- |
- |
hypothetical protein PHAVU_009G146200g [Phaseolus vulgaris] |
| 14 |
Hb_001195_040 |
0.1314364007 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 15 |
Hb_005186_050 |
0.1331349023 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 16 |
Hb_001348_070 |
0.1346159608 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_012508_030 |
0.136693792 |
- |
- |
PREDICTED: uncharacterized protein LOC105636284 [Jatropha curcas] |
| 18 |
Hb_000116_310 |
0.1377960887 |
- |
- |
50S robosomal protein L11, putative [Ricinus communis] |
| 19 |
Hb_000085_240 |
0.1387723894 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 20 |
Hb_001439_150 |
0.1389228412 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |