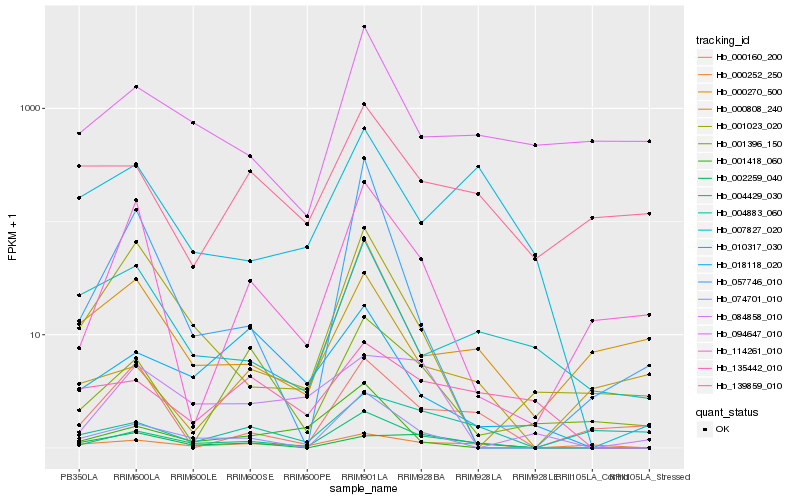
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004429_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s16560g [Populus trichocarpa] |
| 2 |
Hb_074701_010 |
0.1905163203 |
- |
- |
- |
| 3 |
Hb_000252_250 |
0.249428152 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_114261_010 |
0.2773225482 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 5 |
Hb_010317_030 |
0.2866500152 |
- |
- |
unknown [Picea sitchensis] |
| 6 |
Hb_001023_020 |
0.2873463949 |
- |
- |
hypothetical protein POPTR_1605s00200g [Populus trichocarpa] |
| 7 |
Hb_000270_500 |
0.2875984204 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 8 |
Hb_002259_040 |
0.2915414357 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001396_150 |
0.2931213273 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 10 |
Hb_135442_010 |
0.2958921516 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 11 |
Hb_007827_020 |
0.2967970309 |
- |
- |
- |
| 12 |
Hb_001418_060 |
0.2972899032 |
- |
- |
- |
| 13 |
Hb_094647_010 |
0.2983601708 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B-like [Jatropha curcas] |
| 14 |
Hb_057746_010 |
0.298612065 |
- |
- |
- |
| 15 |
Hb_000808_240 |
0.301446126 |
- |
- |
- |
| 16 |
Hb_018118_020 |
0.3022329643 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000160_200 |
0.3060784094 |
- |
- |
PREDICTED: uncharacterized protein LOC105648226 [Jatropha curcas] |
| 18 |
Hb_139859_010 |
0.3080365923 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 19 |
Hb_004883_060 |
0.314916502 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 20 |
Hb_084858_010 |
0.315982316 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |