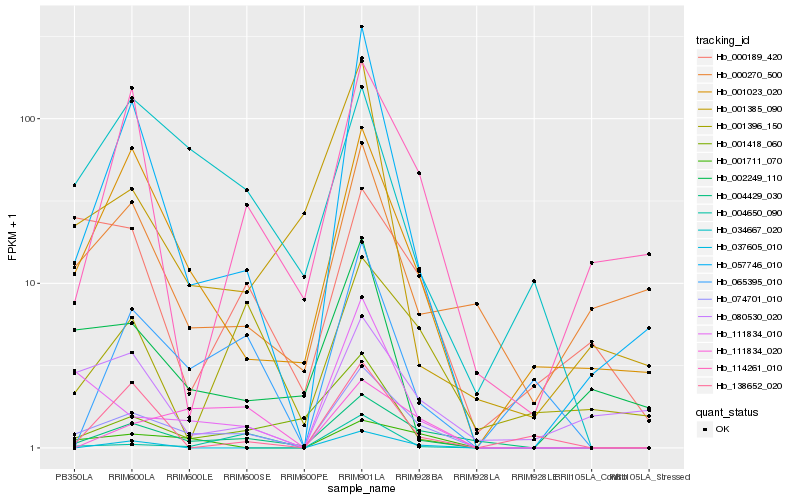
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074701_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004429_030 |
0.1905163203 |
- |
- |
hypothetical protein POPTR_0010s16560g [Populus trichocarpa] |
| 3 |
Hb_057746_010 |
0.232717848 |
- |
- |
- |
| 4 |
Hb_001711_070 |
0.2495509739 |
- |
- |
- |
| 5 |
Hb_111834_010 |
0.2512053619 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 6 |
Hb_001023_020 |
0.2513436513 |
- |
- |
hypothetical protein POPTR_1605s00200g [Populus trichocarpa] |
| 7 |
Hb_065395_010 |
0.279929853 |
- |
- |
hypothetical protein POPTR_0014s14760g, partial [Populus trichocarpa] |
| 8 |
Hb_001418_060 |
0.2842384042 |
- |
- |
- |
| 9 |
Hb_034667_020 |
0.2977993325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_114261_010 |
0.3031865783 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 11 |
Hb_037605_010 |
0.3067746552 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 12 |
Hb_111834_020 |
0.3123130282 |
- |
- |
PREDICTED: protein phosphatase methylesterase 1 [Jatropha curcas] |
| 13 |
Hb_080530_020 |
0.3126002925 |
- |
- |
- |
| 14 |
Hb_004650_090 |
0.3136798176 |
- |
- |
- |
| 15 |
Hb_138652_020 |
0.3167201345 |
- |
- |
- |
| 16 |
Hb_000270_500 |
0.3183902062 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 17 |
Hb_001385_090 |
0.3197574649 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001396_150 |
0.3225065034 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 19 |
Hb_000189_420 |
0.3236255564 |
- |
- |
PREDICTED: uncharacterized protein LOC105647665 [Jatropha curcas] |
| 20 |
Hb_002249_110 |
0.3236418275 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |