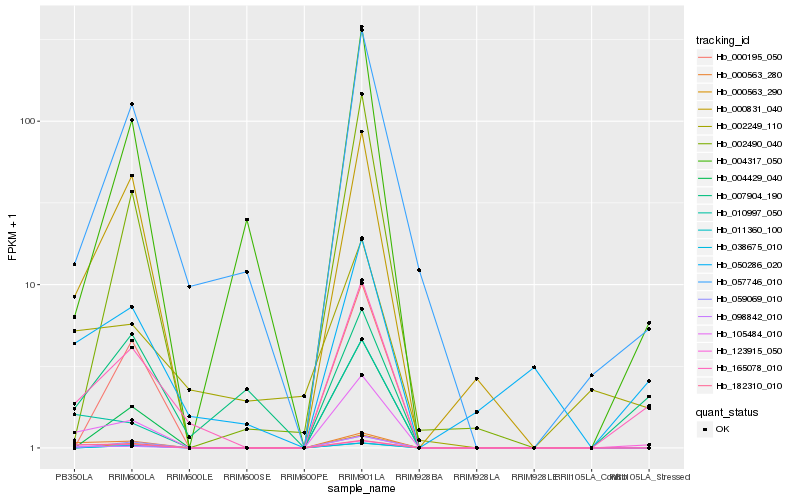
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165078_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_057746_010 |
0.2093928835 |
- |
- |
- |
| 3 |
Hb_105484_010 |
0.215062797 |
- |
- |
carbohydrate-binding-like fold protein, partial [Manihot esculenta] |
| 4 |
Hb_050286_020 |
0.2374515307 |
- |
- |
- |
| 5 |
Hb_000831_040 |
0.2380074061 |
- |
- |
- |
| 6 |
Hb_098842_010 |
0.2554773521 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 7 |
Hb_004317_050 |
0.2560608547 |
- |
- |
- |
| 8 |
Hb_000563_280 |
0.262350483 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 9 |
Hb_007904_190 |
0.2640857931 |
- |
- |
- |
| 10 |
Hb_010997_050 |
0.2711511711 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_000563_290 |
0.2725641524 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 12 |
Hb_002249_110 |
0.273607035 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 13 |
Hb_000195_050 |
0.2744506188 |
- |
- |
- |
| 14 |
Hb_002490_040 |
0.2773987585 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 15 |
Hb_004429_040 |
0.2800709825 |
- |
- |
- |
| 16 |
Hb_182310_010 |
0.2812594926 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 17 |
Hb_038675_010 |
0.2812865409 |
- |
- |
PREDICTED: uncharacterized protein LOC105155307, partial [Sesamum indicum] |
| 18 |
Hb_059069_010 |
0.2815535391 |
- |
- |
PREDICTED: uncharacterized protein LOC105180180, partial [Sesamum indicum] |
| 19 |
Hb_123915_050 |
0.285719315 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 20 |
Hb_011360_100 |
0.2859782268 |
- |
- |
- |