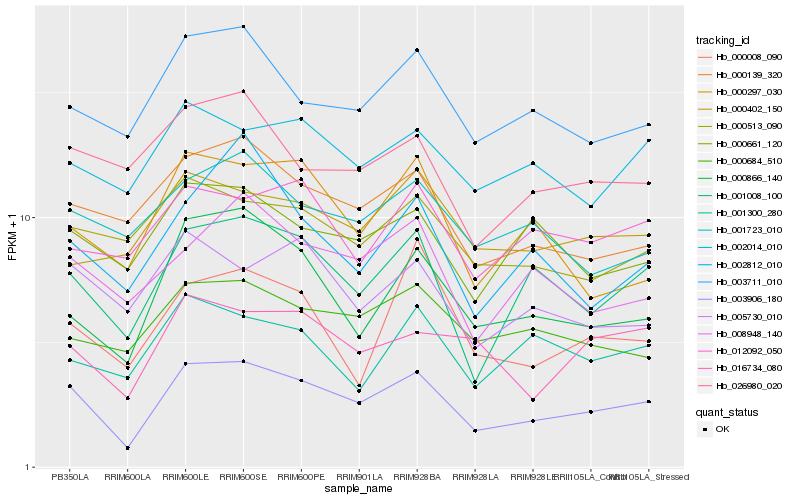
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003906_180 |
0.0 |
- |
- |
unknown [Zea mays] |
| 2 |
Hb_001008_100 |
0.1056806104 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 3 |
Hb_000866_140 |
0.1177297158 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 4 |
Hb_003711_010 |
0.124176784 |
- |
- |
isopropylmalate synthase, putative [Ricinus communis] |
| 5 |
Hb_000139_320 |
0.1277869067 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 6 |
Hb_008948_140 |
0.1301206664 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 7 |
Hb_016734_080 |
0.1346228028 |
- |
- |
PREDICTED: pyridine nucleotide-disulfide oxidoreductase domain-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_000661_120 |
0.1347285486 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 9 |
Hb_000297_030 |
0.1369184897 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000684_510 |
0.137375056 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 11 |
Hb_002014_010 |
0.1378492877 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_005730_010 |
0.1383766885 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 13 |
Hb_002812_010 |
0.1393853118 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_026980_020 |
0.1396928274 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 15 |
Hb_000513_090 |
0.1411773337 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 16 |
Hb_000008_090 |
0.1420102204 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 17 |
Hb_001723_010 |
0.1427964631 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 18 |
Hb_001300_280 |
0.1431455247 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 19 |
Hb_012092_050 |
0.1434072456 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000402_150 |
0.1449760828 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |