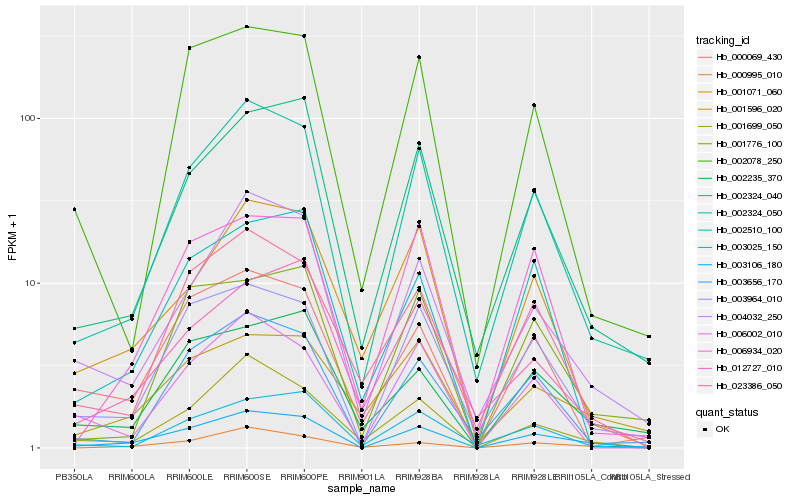
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_180 |
0.0 |
- |
- |
Peroxidase 65 precursor, putative [Ricinus communis] |
| 2 |
Hb_002324_050 |
0.124687988 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 3 |
Hb_003964_010 |
0.1390634536 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 4 |
Hb_004032_250 |
0.1511792209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002324_040 |
0.1527581766 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 6 |
Hb_000995_010 |
0.155373096 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 7 |
Hb_001699_050 |
0.1592687715 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 8 |
Hb_003656_170 |
0.1617208024 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 9 |
Hb_002235_370 |
0.1661480466 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 10 |
Hb_023386_050 |
0.1666114203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000069_430 |
0.1709407383 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 12 |
Hb_002510_100 |
0.1725028998 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 13 |
Hb_001071_060 |
0.1725523886 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 14 |
Hb_002078_250 |
0.1738775325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003025_150 |
0.1759814342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001776_100 |
0.176712631 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006934_020 |
0.1784878228 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 18 |
Hb_006002_010 |
0.1793451445 |
transcription factor |
TF Family: MYB |
Myb domain protein 5 isoform 1 [Theobroma cacao] |
| 19 |
Hb_012727_010 |
0.1813886637 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 20 |
Hb_001596_020 |
0.1825209566 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |