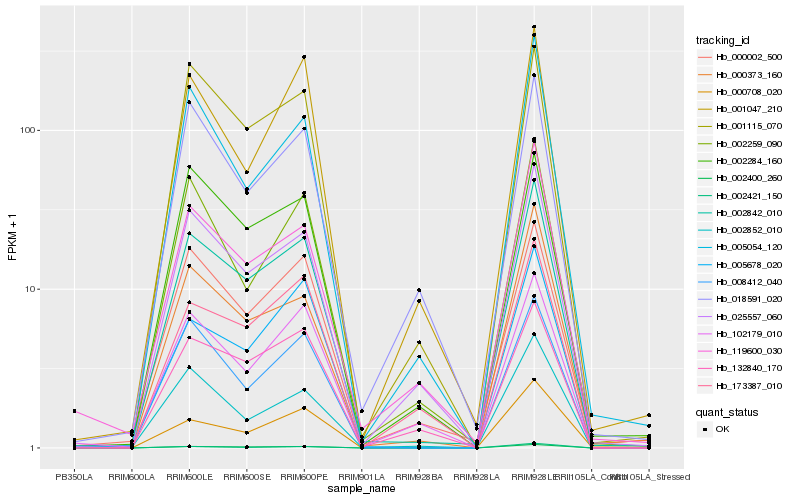
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002842_010 |
0.0 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 2 |
Hb_002400_260 |
0.0524814898 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_025557_060 |
0.0798664721 |
transcription factor |
TF Family: MYB |
Myb-related protein 306, putative isoform 1 [Theobroma cacao] |
| 4 |
Hb_000373_160 |
0.0926874949 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 5 |
Hb_002421_150 |
0.0946552464 |
- |
- |
hypothetical protein PRUPE_ppa020511mg [Prunus persica] |
| 6 |
Hb_000002_500 |
0.0969066884 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 7 |
Hb_119600_030 |
0.0980316201 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 8 |
Hb_002852_010 |
0.1004511118 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_000708_020 |
0.1009654903 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_001115_070 |
0.1016853438 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 11 |
Hb_005054_120 |
0.1018862278 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008412_040 |
0.1022534515 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_002259_090 |
0.1068274717 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 14 |
Hb_005678_020 |
0.1091149819 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002284_160 |
0.1120479334 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_102179_010 |
0.1139087997 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_132840_170 |
0.113943622 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 18 |
Hb_173387_010 |
0.1141305463 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 19 |
Hb_018591_020 |
0.1146093531 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001047_210 |
0.1204627107 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |