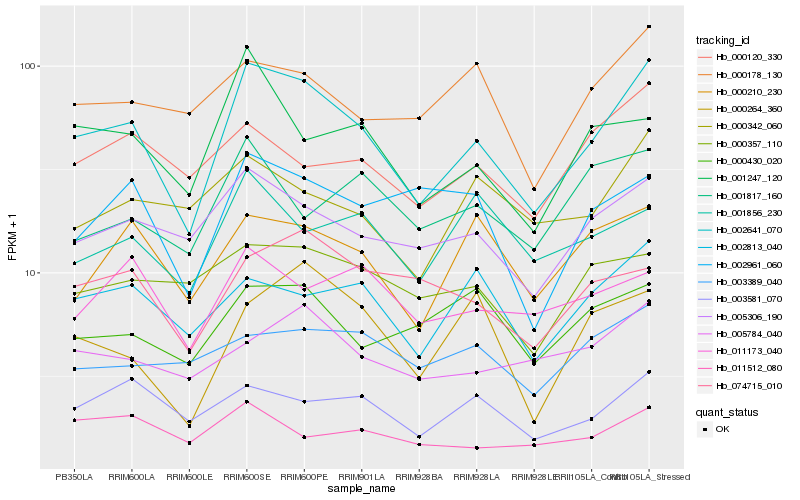
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002641_070 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 2 |
Hb_005306_190 |
0.12973667 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 3 |
Hb_002813_040 |
0.1444491682 |
- |
- |
PREDICTED: PRA1 family protein G2 [Jatropha curcas] |
| 4 |
Hb_003581_070 |
0.1445464027 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Malus domestica] |
| 5 |
Hb_000210_230 |
0.1469514035 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 6 |
Hb_001247_120 |
0.1488535295 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 7 |
Hb_000178_130 |
0.1504556142 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 8 |
Hb_000120_330 |
0.1535354326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005784_040 |
0.154069577 |
- |
- |
PREDICTED: uncharacterized protein LOC105645160 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000342_060 |
0.1545769541 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |
| 11 |
Hb_074715_010 |
0.1555777633 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002961_060 |
0.1567543847 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 5 [Populus euphratica] |
| 13 |
Hb_000430_020 |
0.1579320468 |
- |
- |
- |
| 14 |
Hb_000264_360 |
0.1600070547 |
- |
- |
hypothetical protein JCGZ_21443 [Jatropha curcas] |
| 15 |
Hb_011173_040 |
0.160538785 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Populus euphratica] |
| 16 |
Hb_001856_230 |
0.161532099 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 17 |
Hb_003389_040 |
0.161692082 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 18 |
Hb_000357_110 |
0.1618056459 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 19 |
Hb_011512_080 |
0.1618721846 |
- |
- |
hypothetical protein JCGZ_06424 [Jatropha curcas] |
| 20 |
Hb_001817_160 |
0.1635566253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |