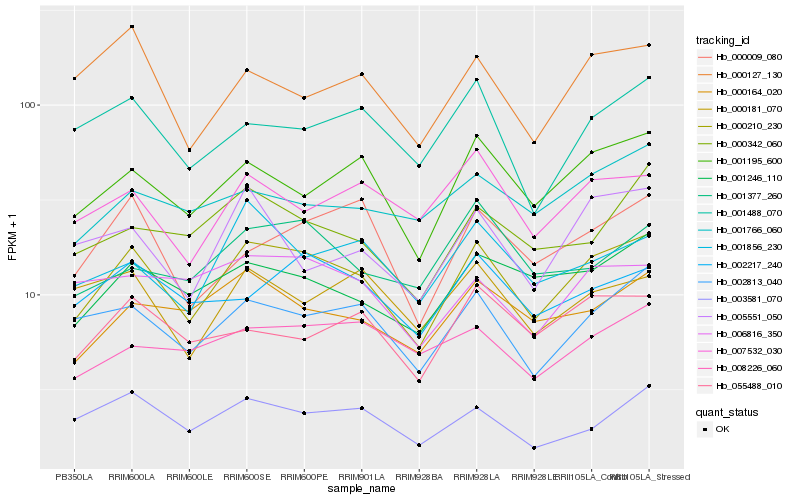
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 2 |
Hb_001195_600 |
0.0978699045 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 3 |
Hb_002813_040 |
0.0999339007 |
- |
- |
PREDICTED: PRA1 family protein G2 [Jatropha curcas] |
| 4 |
Hb_001246_110 |
0.1067344858 |
- |
- |
PREDICTED: probable purine permease 11 [Jatropha curcas] |
| 5 |
Hb_000164_020 |
0.1097296634 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 6 |
Hb_003581_070 |
0.1109048495 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Malus domestica] |
| 7 |
Hb_001856_230 |
0.1166107674 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 8 |
Hb_002217_240 |
0.1180886674 |
- |
- |
PREDICTED: F-box protein At5g49610 [Jatropha curcas] |
| 9 |
Hb_007532_030 |
0.1181407423 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 10 |
Hb_001488_070 |
0.1187962945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000342_060 |
0.1192354044 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like isoform X1 [Nicotiana sylvestris] |
| 12 |
Hb_000009_080 |
0.1193945593 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 15 [Jatropha curcas] |
| 13 |
Hb_055488_010 |
0.1203620425 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000127_130 |
0.1210707524 |
- |
- |
PREDICTED: uncharacterized protein LOC105635483 [Jatropha curcas] |
| 15 |
Hb_001766_060 |
0.1240027315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005551_050 |
0.125883222 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 17 |
Hb_008226_060 |
0.1262078375 |
- |
- |
PREDICTED: folic acid synthesis protein fol1-like [Jatropha curcas] |
| 18 |
Hb_006816_350 |
0.1262315691 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 19 |
Hb_001377_260 |
0.1263676709 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000181_070 |
0.1268249389 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |