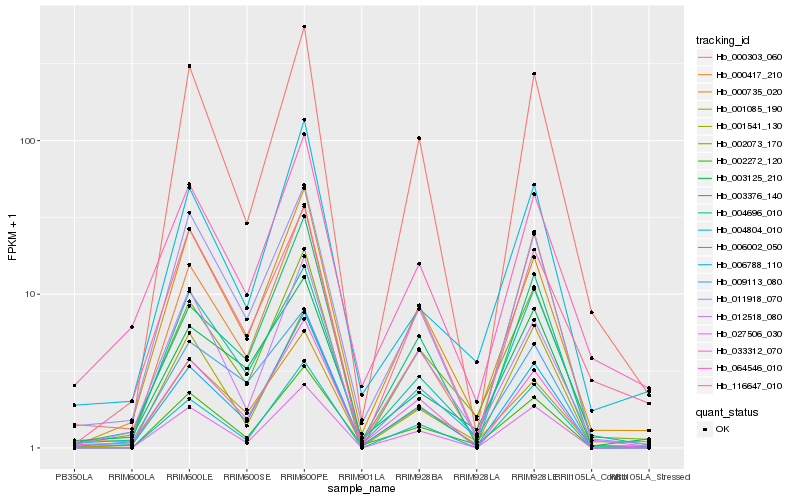
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_170 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g27190 [Jatropha curcas] |
| 2 |
Hb_006788_110 |
0.1063116889 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 3 |
Hb_000735_020 |
0.1089788063 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 4 |
Hb_004696_010 |
0.1105639152 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 5 |
Hb_000303_060 |
0.1161329703 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003125_210 |
0.1163393737 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 7 |
Hb_001085_190 |
0.1179758982 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_000417_210 |
0.1184232457 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_002272_120 |
0.1217421525 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 10 |
Hb_012518_080 |
0.1243889925 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 11 |
Hb_064546_010 |
0.1261641673 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 12 |
Hb_004804_010 |
0.1284878683 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 13 |
Hb_003376_140 |
0.1296279094 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 14 |
Hb_009113_080 |
0.1296703766 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 15 |
Hb_033312_070 |
0.1296811021 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 16 |
Hb_027506_030 |
0.1307792589 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_011918_070 |
0.1310040744 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 18 |
Hb_006002_050 |
0.1318645957 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 19 |
Hb_001541_130 |
0.1338062087 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 20 |
Hb_116647_010 |
0.1340692409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |