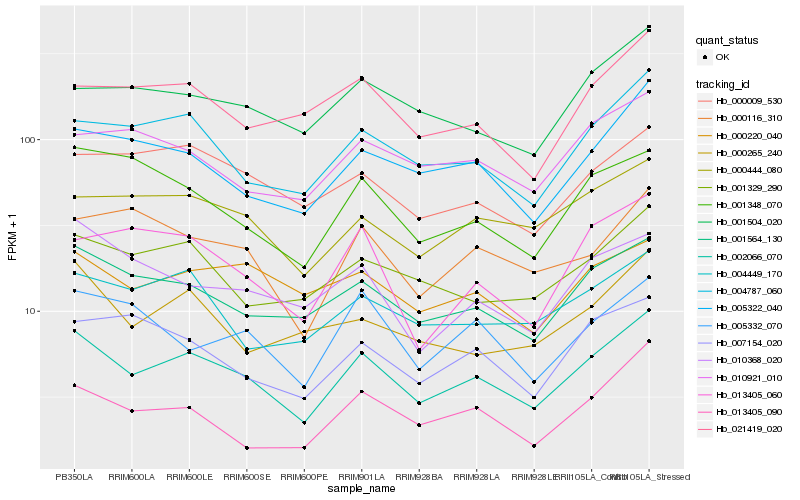
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002066_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631043 [Jatropha curcas] |
| 2 |
Hb_001564_130 |
0.0899463558 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_004787_060 |
0.0921052011 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 4 |
Hb_005322_040 |
0.0968958677 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 5 |
Hb_001329_290 |
0.1065864683 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 6 |
Hb_000444_080 |
0.1073591793 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 7 |
Hb_004449_170 |
0.1114288111 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001348_070 |
0.1124038388 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000009_530 |
0.1134152709 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 10 |
Hb_007154_020 |
0.1135448765 |
- |
- |
- |
| 11 |
Hb_001504_020 |
0.11378686 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 12 |
Hb_013405_090 |
0.1142196315 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000116_310 |
0.1148674505 |
- |
- |
50S robosomal protein L11, putative [Ricinus communis] |
| 14 |
Hb_021419_020 |
0.1155249237 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 15 |
Hb_010921_010 |
0.1165490216 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 16 |
Hb_000265_240 |
0.1168095781 |
- |
- |
PREDICTED: uncharacterized protein LOC105643590 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000220_040 |
0.1172896421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_013405_060 |
0.1174849914 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005332_070 |
0.1178771911 |
- |
- |
PREDICTED: uncharacterized protein LOC105632487 [Jatropha curcas] |
| 20 |
Hb_010368_020 |
0.1186217623 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |