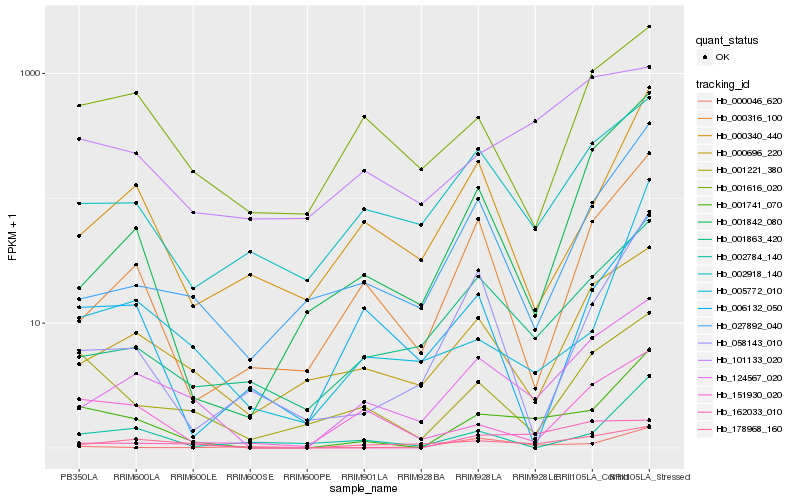
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001741_070 |
0.0 |
rubber biosynthesis |
Gene Name: SRPP7 |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_124567_020 |
0.2412628399 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 3 |
Hb_001221_380 |
0.252753173 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 4 |
Hb_005772_010 |
0.2688662266 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 5 |
Hb_001863_420 |
0.2693786715 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 6 |
Hb_151930_020 |
0.2700549483 |
- |
- |
Wound-induced protein WIN1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002784_140 |
0.2775580089 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 8 |
Hb_002918_140 |
0.2778530974 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 9 |
Hb_000340_440 |
0.2807024514 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 10 |
Hb_178968_160 |
0.2850685716 |
- |
- |
- |
| 11 |
Hb_001842_080 |
0.2875159733 |
- |
- |
- |
| 12 |
Hb_027892_040 |
0.288908011 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 13 |
Hb_001616_020 |
0.2892317653 |
- |
- |
PREDICTED: wound-induced basic protein [Jatropha curcas] |
| 14 |
Hb_000316_100 |
0.2901922684 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_000696_220 |
0.290871557 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 16 |
Hb_006132_050 |
0.2917092793 |
- |
- |
hypothetical protein EUTSA_v10028860mg [Eutrema salsugineum] |
| 17 |
Hb_058143_010 |
0.2950746306 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 18 |
Hb_000046_620 |
0.2957264509 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 19 |
Hb_101133_020 |
0.2995671192 |
- |
- |
PREDICTED: 15.7 kDa heat shock protein, peroxisomal [Jatropha curcas] |
| 20 |
Hb_162033_010 |
0.3025853983 |
- |
- |
- |