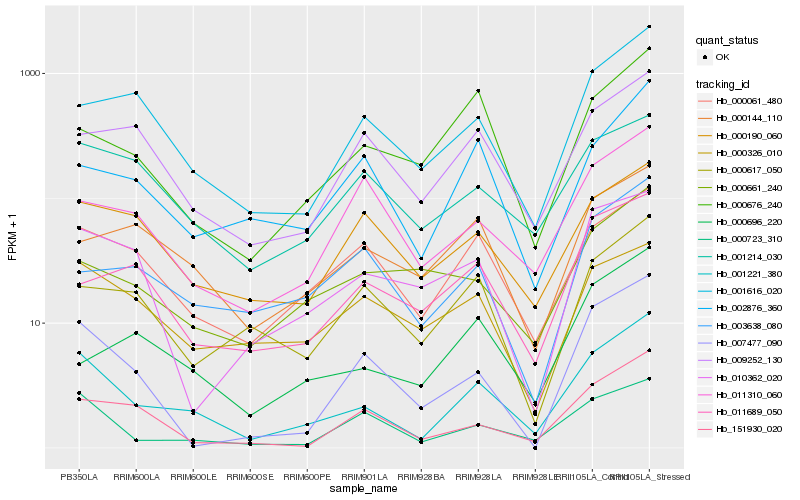
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_380 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 2 |
Hb_000723_310 |
0.1703170227 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_001616_020 |
0.1813270777 |
- |
- |
PREDICTED: wound-induced basic protein [Jatropha curcas] |
| 4 |
Hb_000061_480 |
0.1881910263 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 5 |
Hb_151930_020 |
0.1888910867 |
- |
- |
Wound-induced protein WIN1 precursor, putative [Ricinus communis] |
| 6 |
Hb_007477_090 |
0.1920977406 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 7 |
Hb_002876_360 |
0.1971710729 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 8 |
Hb_000676_240 |
0.1982797005 |
- |
- |
thioredoxin H-type 2 [Hevea brasiliensis] |
| 9 |
Hb_001214_030 |
0.2046671319 |
- |
- |
hypothetical protein POPTR_0006s10280g [Populus trichocarpa] |
| 10 |
Hb_009252_130 |
0.2053295825 |
- |
- |
Deletion of SUV3 suppressor 1(I) isoform 1 [Theobroma cacao] |
| 11 |
Hb_000190_060 |
0.2074986104 |
- |
- |
PREDICTED: uncharacterized protein LOC105644688 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000696_220 |
0.2085833991 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 13 |
Hb_000144_110 |
0.2096355634 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 14 |
Hb_011310_060 |
0.2102397433 |
- |
- |
PREDICTED: uncharacterized protein At2g34160-like [Jatropha curcas] |
| 15 |
Hb_011689_050 |
0.2104852752 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 16 |
Hb_003638_080 |
0.2118152335 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 17 |
Hb_000617_050 |
0.2124073854 |
- |
- |
PREDICTED: uncharacterized protein LOC105647466 [Jatropha curcas] |
| 18 |
Hb_010362_020 |
0.2163222893 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 38-like [Jatropha curcas] |
| 19 |
Hb_000661_240 |
0.2176181583 |
- |
- |
hypothetical protein POPTR_0009s05540g [Populus trichocarpa] |
| 20 |
Hb_000326_010 |
0.2176451711 |
- |
- |
hypothetical protein POPTR_0009s11870g [Populus trichocarpa] |