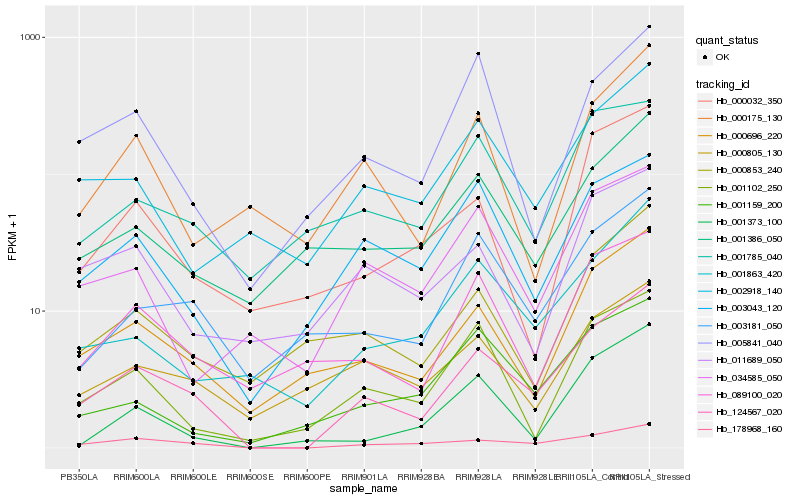
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124567_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 2 |
Hb_178968_160 |
0.1255283679 |
- |
- |
- |
| 3 |
Hb_000696_220 |
0.1575554775 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 4 |
Hb_001863_420 |
0.174188296 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 5 |
Hb_003181_050 |
0.1786621121 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 6 |
Hb_089100_020 |
0.185963021 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 7 |
Hb_002918_140 |
0.1870019729 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 8 |
Hb_003043_120 |
0.1876447969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000853_240 |
0.1885997172 |
- |
- |
- |
| 10 |
Hb_001386_050 |
0.1929207256 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 11 |
Hb_001102_250 |
0.1950111083 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001159_200 |
0.1991054059 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_034585_050 |
0.1992669312 |
- |
- |
- |
| 14 |
Hb_005841_040 |
0.1994522088 |
- |
- |
stress-induced hydrophobic peptide 1 [Hevea brasiliensis] |
| 15 |
Hb_000175_130 |
0.1996508911 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 16 |
Hb_000032_350 |
0.2003945433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000805_130 |
0.2010141545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011689_050 |
0.2032514512 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 19 |
Hb_001373_100 |
0.2055426618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001785_040 |
0.206433067 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |