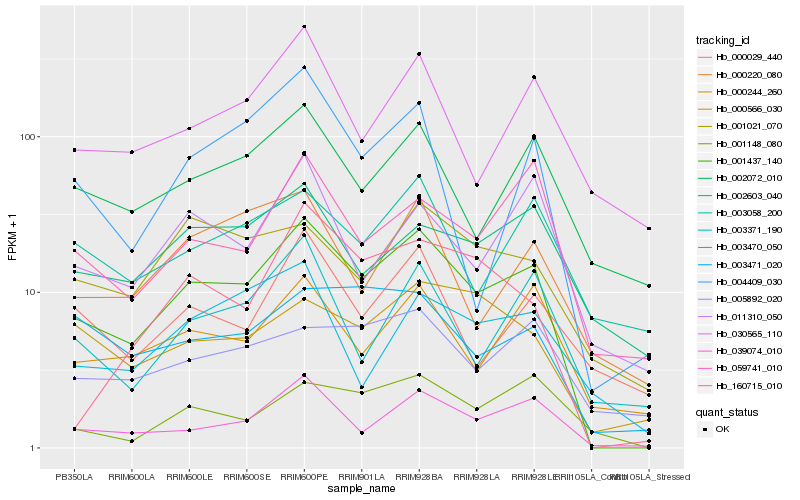
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_039074_010 |
0.1559163743 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_000244_260 |
0.178394605 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002072_010 |
0.1810382661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_059741_010 |
0.1873236645 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 6 |
Hb_004409_030 |
0.1884375637 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |
| 7 |
Hb_003471_020 |
0.1887833936 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Jatropha curcas] |
| 8 |
Hb_003371_190 |
0.1920825832 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 9 |
Hb_030565_110 |
0.1921821799 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 10 |
Hb_001148_080 |
0.1926834262 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 11 |
Hb_000029_440 |
0.1944149239 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |
| 12 |
Hb_001021_070 |
0.1956920578 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_003058_200 |
0.1957472311 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 14 |
Hb_000566_030 |
0.1957539859 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 15 |
Hb_160715_010 |
0.196159298 |
- |
- |
- |
| 16 |
Hb_005892_020 |
0.1961596022 |
- |
- |
PREDICTED: uncharacterized protein LOC105632170 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011310_050 |
0.1964316267 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 18 |
Hb_002603_040 |
0.1969653693 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |
| 19 |
Hb_000220_080 |
0.1971168392 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_003470_050 |
0.2026244598 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |