| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
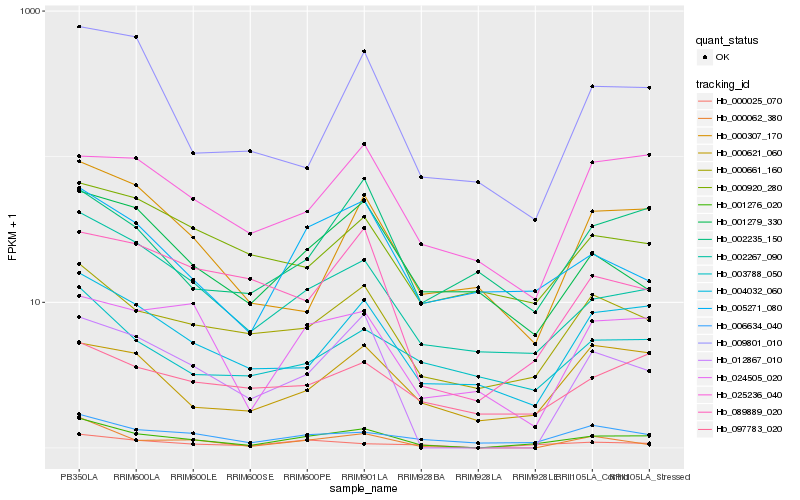
Hb_001276_020 |
0.0 |
- |
- |
hypothetical protein VITISV_003263 [Vitis vinifera] |
| 2 |
Hb_002267_090 |
0.1596128872 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 3 |
Hb_000661_160 |
0.1650921981 |
- |
- |
hexokinase [Manihot esculenta] |
| 4 |
Hb_000025_070 |
0.1701755113 |
- |
- |
Uncharacterized protein TCM_040985 [Theobroma cacao] |
| 5 |
Hb_004032_060 |
0.1795183494 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |
| 6 |
Hb_006634_040 |
0.1821123131 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 7 |
Hb_005271_080 |
0.1823793276 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 8 |
Hb_000621_060 |
0.1991316927 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 9 |
Hb_089889_020 |
0.2014720851 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 10 |
Hb_012867_010 |
0.2035234272 |
- |
- |
Vacuolar protein sorting 45 [Theobroma cacao] |
| 11 |
Hb_000062_380 |
0.2041872058 |
- |
- |
PREDICTED: putative F-box protein At1g67623 [Jatropha curcas] |
| 12 |
Hb_001279_330 |
0.2042782488 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 13 |
Hb_003788_050 |
0.2064717596 |
- |
- |
PREDICTED: interferon-induced guanylate-binding protein 2-like [Jatropha curcas] |
| 14 |
Hb_097783_020 |
0.2102014613 |
- |
- |
PREDICTED: small G protein signaling modulator 1 [Jatropha curcas] |
| 15 |
Hb_024505_020 |
0.2113963984 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 16 |
Hb_000307_170 |
0.2121065838 |
- |
- |
PREDICTED: uncharacterized protein LOC105638528 [Jatropha curcas] |
| 17 |
Hb_002235_150 |
0.213114431 |
- |
- |
- |
| 18 |
Hb_009801_010 |
0.2133431583 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 19 |
Hb_000920_280 |
0.2136092337 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 20 |
Hb_025236_040 |
0.2139535698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |