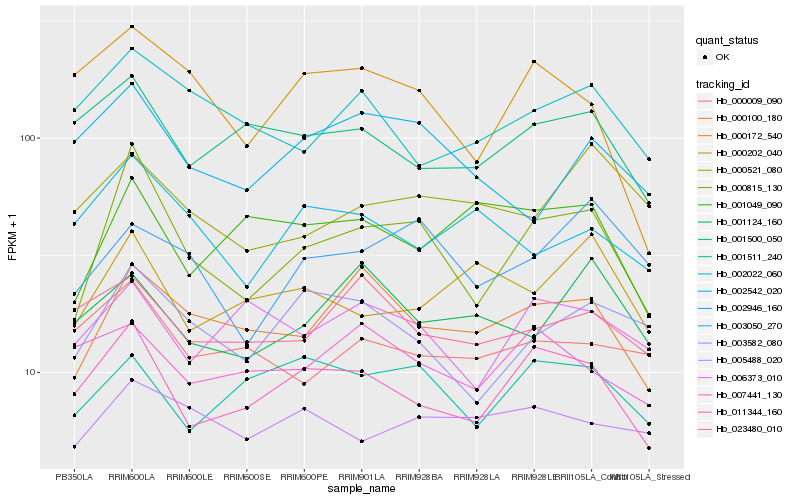
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 2 |
Hb_011344_160 |
0.1255827288 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 3 |
Hb_000100_180 |
0.1328792398 |
- |
- |
PREDICTED: probable acyl-activating enzyme 16, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003582_080 |
0.1451427744 |
- |
- |
iaa-alanine resistance protein, putative [Ricinus communis] |
| 5 |
Hb_003050_270 |
0.1574860971 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001511_240 |
0.1603142197 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 7 |
Hb_000172_540 |
0.1613151288 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 8 |
Hb_006373_010 |
0.1639066792 |
- |
- |
Potassium transporter 11 [Gossypium arboreum] |
| 9 |
Hb_001500_050 |
0.1660404333 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 10 |
Hb_002022_060 |
0.1673027903 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 11 |
Hb_002946_160 |
0.1677950327 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 12 |
Hb_005488_020 |
0.1701584175 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 13 |
Hb_000521_080 |
0.1704606599 |
- |
- |
Zinc finger BED domain-containing 4 [Gossypium arboreum] |
| 14 |
Hb_001049_090 |
0.1714762616 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_023480_010 |
0.1723310356 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000009_090 |
0.173867215 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 17 |
Hb_000202_040 |
0.1739631422 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 18 |
Hb_002542_020 |
0.1750048255 |
- |
- |
PREDICTED: uncharacterized protein LOC105634003 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001124_160 |
0.1767177195 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 20 |
Hb_007441_130 |
0.1768723214 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |