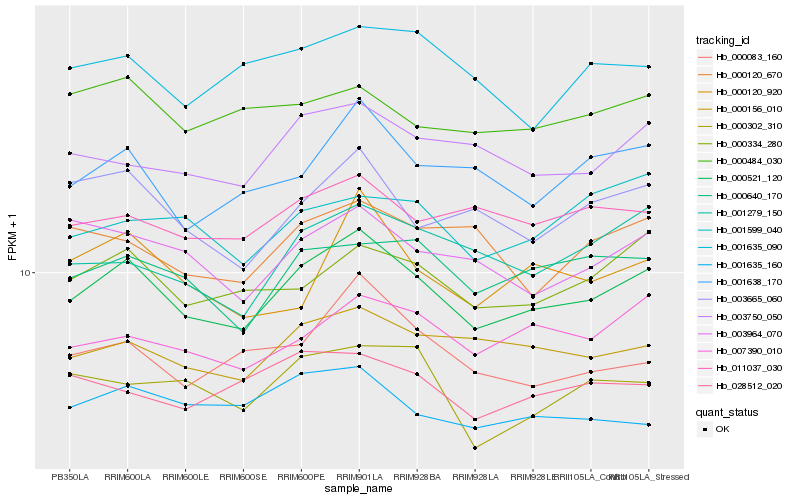
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_120 |
0.0 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_000156_010 |
0.0720442419 |
- |
- |
phospho-n-acetylmuramoyl-pentapeptide-transferase, putative [Ricinus communis] |
| 3 |
Hb_000334_280 |
0.072229184 |
- |
- |
PREDICTED: protein FRA10AC1 [Jatropha curcas] |
| 4 |
Hb_001638_170 |
0.0740977739 |
- |
- |
hypothetical protein Csa_3G118170 [Cucumis sativus] |
| 5 |
Hb_000640_170 |
0.074433531 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007390_010 |
0.0753381367 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_003750_050 |
0.0765299317 |
- |
- |
PREDICTED: THO complex subunit 4D isoform X1 [Jatropha curcas] |
| 8 |
Hb_003665_060 |
0.0766489517 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 9 |
Hb_003964_070 |
0.0783585664 |
- |
- |
hypothetical protein JCGZ_15695 [Jatropha curcas] |
| 10 |
Hb_001279_150 |
0.0799132285 |
- |
- |
PREDICTED: ATP synthase mitochondrial F1 complex assembly factor 2 [Jatropha curcas] |
| 11 |
Hb_000083_160 |
0.0806509296 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 12 |
Hb_011037_030 |
0.0820451676 |
- |
- |
PREDICTED: uncharacterized protein LOC105641407 [Jatropha curcas] |
| 13 |
Hb_001635_160 |
0.0839730294 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 14 |
Hb_000120_920 |
0.0855663081 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 15 |
Hb_000484_030 |
0.0858054889 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 16 |
Hb_028512_020 |
0.086152484 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001635_090 |
0.086219704 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 18 |
Hb_000120_670 |
0.0872638728 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 19 |
Hb_000302_310 |
0.0879528196 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 20 |
Hb_001599_040 |
0.0885569298 |
- |
- |
PREDICTED: suppressor protein SRP40 isoform X2 [Jatropha curcas] |