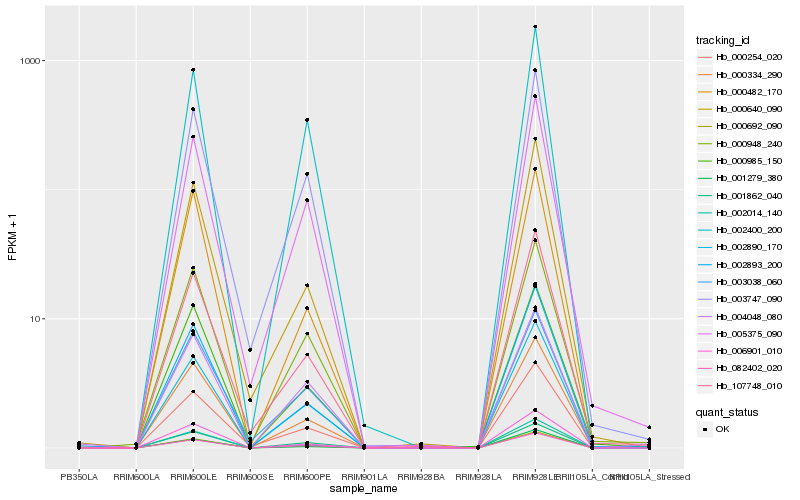
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_004048_080 |
0.1069922688 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 3 |
Hb_082402_020 |
0.1097410234 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 4 |
Hb_002890_170 |
0.1115021719 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 5 |
Hb_002014_140 |
0.1126781906 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 6 |
Hb_000334_290 |
0.115177391 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 7 |
Hb_006901_010 |
0.1162968654 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 8 |
Hb_000254_020 |
0.1170646712 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 9 |
Hb_003038_060 |
0.1185297084 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 10 |
Hb_001862_040 |
0.119378491 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 11 |
Hb_002893_200 |
0.1215707798 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_003747_090 |
0.1220431309 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_005375_090 |
0.1232131226 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000640_090 |
0.1247279275 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000985_150 |
0.1247833947 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 16 |
Hb_001279_380 |
0.1260344473 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 17 |
Hb_000948_240 |
0.1261130874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000482_170 |
0.1261559593 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 19 |
Hb_107748_010 |
0.1262310991 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_002400_200 |
0.1279179896 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |