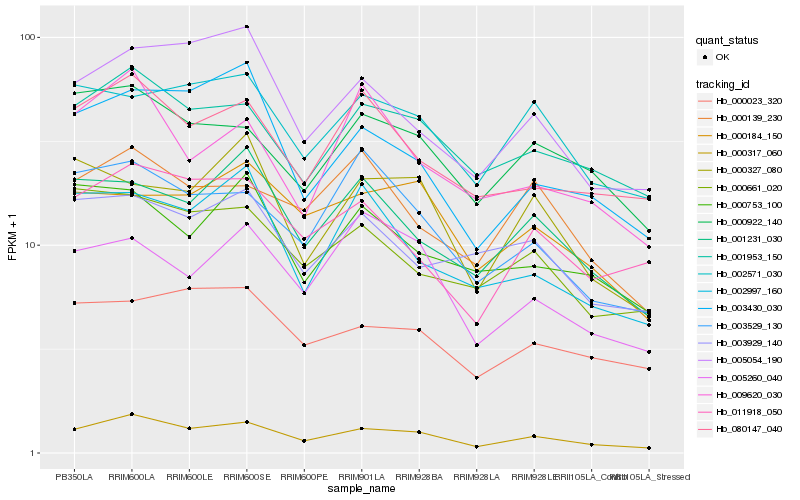
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647051 [Jatropha curcas] |
| 2 |
Hb_001953_150 |
0.0934815112 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 3 |
Hb_005054_190 |
0.1065789632 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 4 |
Hb_080147_040 |
0.106756328 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 5 |
Hb_000922_140 |
0.106939306 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 6 |
Hb_003430_030 |
0.1090919335 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 7 |
Hb_011918_050 |
0.110135088 |
- |
- |
- |
| 8 |
Hb_003529_130 |
0.1118894025 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_001231_030 |
0.114189841 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 10 |
Hb_002571_030 |
0.1178191788 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 11 |
Hb_000661_020 |
0.1184543432 |
- |
- |
BnaC08g27190D [Brassica napus] |
| 12 |
Hb_000023_320 |
0.1214839561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000139_230 |
0.1216984432 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 14 |
Hb_005260_040 |
0.1232556108 |
- |
- |
PREDICTED: uncharacterized protein LOC105633768 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002997_160 |
0.1244159386 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_009620_030 |
0.1257331101 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 17 |
Hb_003929_140 |
0.1259154332 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000184_150 |
0.1260540728 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 19 |
Hb_000327_080 |
0.1264500695 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 20 |
Hb_000753_100 |
0.1268344938 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |