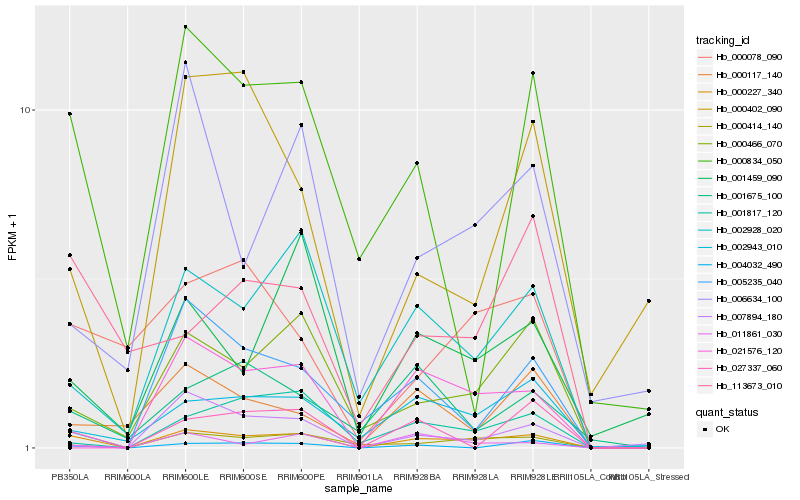
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_340 |
0.0 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 2 |
Hb_000466_070 |
0.2232051153 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 3 |
Hb_002928_020 |
0.2240935785 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 4 |
Hb_000414_140 |
0.2319760106 |
- |
- |
Chalcone synthase, putative [Ricinus communis] |
| 5 |
Hb_002943_010 |
0.237199252 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 6 |
Hb_011861_030 |
0.2542195875 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance protein [Morus notabilis] |
| 7 |
Hb_000078_090 |
0.2619195702 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 8 |
Hb_000834_050 |
0.2625794022 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 9 |
Hb_001459_090 |
0.2633942111 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000117_140 |
0.2667314471 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |
| 11 |
Hb_001817_120 |
0.2676007944 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_027337_060 |
0.2676968673 |
- |
- |
- |
| 13 |
Hb_004032_490 |
0.2681958299 |
- |
- |
PREDICTED: uncharacterized protein LOC100853401 [Vitis vinifera] |
| 14 |
Hb_006634_100 |
0.2726835037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_113673_010 |
0.2730067283 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 16 |
Hb_001675_100 |
0.2799419897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000402_090 |
0.2818484761 |
- |
- |
Lipase [Glycine soja] |
| 18 |
Hb_005235_040 |
0.2822375846 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 19 |
Hb_021576_120 |
0.2852255947 |
- |
- |
DNA-directed RNA polymerase subunit, putative [Ricinus communis] |
| 20 |
Hb_007894_180 |
0.2888357201 |
- |
- |
PREDICTED: cytochrome P450 716B1-like [Jatropha curcas] |